For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAFVLVCPDALAIAAGQLRHVGSVIAARNAVAAPATAELAPAAADEVSALTATQFNFHAAMYQAVGAQAI AMNEAFVAMLGASADSYAATEAANIIAVS
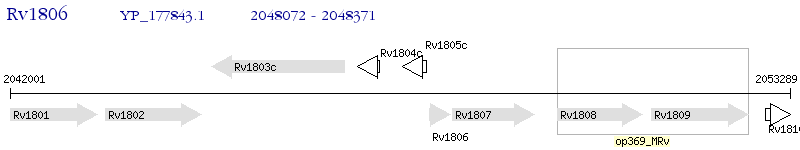
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1806 | PE20 | - | 100% (99) | PE FAMILY PROTEIN |
| M. tuberculosis H37Rv | Rv1788 | PE18 | 1e-24 | 59.79% (97) | PE family protein |
| M. tuberculosis H37Rv | Rv1791 | PE19 | 5e-24 | 58.76% (97) | PE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1835 | PE20 | 2e-49 | 100.00% (99) | PE family protein |
| M. gilvum PYR-GCK | Mflv_0326 | - | 2e-06 | 31.03% (87) | PE domain-containing protein |
| M. leprae Br4923 | MLBr_01183 | - | 5e-13 | 41.38% (87) | hypothetical protein MLBr_01183 |
| M. abscessus ATCC 19977 | MAB_2231c | - | 2e-05 | 32.94% (85) | PE family protein |
| M. marinum M | MMAR_4611 | - | 9e-25 | 54.64% (97) | PE family protein |
| M. avium 104 | MAV_1346 | - | 3e-20 | 50.51% (99) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0618 | - | 3e-07 | 34.48% (87) | PE family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4462 | - | 6e-25 | 54.64% (97) | PE family protein |
| M. vanbaalenii PYR-1 | Mvan_0414 | - | 2e-08 | 36.78% (87) | PE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2231c|M.abscessus_ATCC_199 MN--LNVVPEGLTATSAAVEALTARLAAAHAAAAPVIGAVVPPAADPVSL
MSMEG_0618|M.smegmatis_MC2_155 MT--LRVVPEGLTAASSAVEALTARLAAAHAAAAPMISTVLPPAADAVSL
Mflv_0326|M.gilvum_PYR-GCK MT--LRVVPEGLTAASAAVEALTARMAAAHAAAAPLVTAVLPPAADAVSL
Mvan_0414|M.vanbaalenii_PYR-1 MT--LRVVPEGLTAAGAAVEALTARMAAAHAAAAPVVTAVLPPAADVVSL
Rv1806|M.tuberculosis_H37Rv MA-FVLVCPDALAIAAGQLRHVGSVIAARNAVAAPATAELAPAAADEVSA
Mb1835|M.bovis_AF2122/97 MA-FVLVCPDALAIAAGQLRHVGSVIAARNAVAAPATAELAPAAADEVSA
MMAR_4611|M.marinum_M MS-FVVTQPERLAAAAGSLQEVGSAVSAQSAAAVAPTTSVAPAAADQVSA
MUL_4462|M.ulcerans_Agy99 MS-FVVTQPERLAAAAGSLQEVGSAVSAQSAAAVAPTTSVAPAAADQVSA
MAV_1346|M.avium_104 MS-FVTTRPEALLAVASMLEGLGSSMDAQNAAAAAPTTSIAPAAADEVSA
MLBr_01183|M.leprae_Br4923 MPLFLNAEPQALTAAANTLEGLSAATVASNAAAAQLTTEIAPPAADDVSI
* : . *: * .. :. : : * *.*. : *.*** **
MAB_2231c|M.abscessus_ATCC_199 ETAAGFSARGIEHSGVAAQAVEELGRAGLGVSESAASYTTGDMQAAAA--
MSMEG_0618|M.smegmatis_MC2_155 QTAAGFSANGAQQSAVAAQGVEELGRSGVGVGESGVSYATGDAQAAAS--
Mflv_0326|M.gilvum_PYR-GCK QTATGLSANGVEHQSIAARGVEELGRSGVGVGDSAASYVSGDAMAATS--
Mvan_0414|M.vanbaalenii_PYR-1 QAATGFSARAAEHQSVAAQGVEELGRSGIAVGESAASYAGGDAMAASS--
Rv1806|M.tuberculosis_H37Rv LTATQFNFHAAMYQAVGAQAIAMNEAFVAMLGASADSYAATEAAN-----
Mb1835|M.bovis_AF2122/97 LTATQFNFHAAMYQAVGAQAIAMNEAFVAMLGASADSYAATEAAN-----
MMAR_4611|M.marinum_M LTAARFSAHAQLYQMVSAQAVAIHDRFVAALAAGAQSYEAAEAAN-----
MUL_4462|M.ulcerans_Agy99 LTAARFSAHAQLYQMVSAQAVAIHDRFVAALAAGAQSYEAAEAAN-----
MAV_1346|M.avium_104 LQAAQFSAYGTWYQQVSAQAKAIHQTLVQNLDTNADSYGATEAANRVNTS
MLBr_01183|M.leprae_Br4923 LLAHFFSGHGRQYQAHASQGATNHQDLIQSLLTSSSAYAGTETAN-----
* :. . . .::. : .. :* :
MAB_2231c|M.abscessus_ATCC_199 ---------YMIARG-----------------------------------
MSMEG_0618|M.smegmatis_MC2_155 ---------YLTARGL----------------------------------
Mflv_0326|M.gilvum_PYR-GCK ---------YLIARG-----------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 ---------YLIARG-----------------------------------
Rv1806|M.tuberculosis_H37Rv ----------IIA----------VS-------------------------
Mb1835|M.bovis_AF2122/97 ----------IIA----------VS-------------------------
MMAR_4611|M.marinum_M ----------VIA----------AG-------------------------
MUL_4462|M.ulcerans_Agy99 ----------VIA----------AG-------------------------
MAV_1346|M.avium_104 AAALQGLAPNVIPNAAPAADPPPAGTTSLGTSIGWAQNVGAAASDFITLG
MLBr_01183|M.leprae_Br4923 --------HDSL--------------------------------------
MAB_2231c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1806|M.tuberculosis_H37Rv --------------------------------------------------
Mb1835|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4611|M.marinum_M --------------------------------------------------
MUL_4462|M.ulcerans_Agy99 --------------------------------------------------
MAV_1346|M.avium_104 EGQFAPVPGAAGIPNFNPGLTAATTAGPPAPAAPAAAAGAPVLASMGRGP
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAB_2231c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1806|M.tuberculosis_H37Rv --------------------------------------------------
Mb1835|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4611|M.marinum_M --------------------------------------------------
MUL_4462|M.ulcerans_Agy99 --------------------------------------------------
MAV_1346|M.avium_104 SIGGLSVPPSWAPGGVAPAANATPAQLVGAGFTSAAPDTAPVTAVPGGVP
MLBr_01183|M.leprae_Br4923 --------------------------------------------------
MAB_2231c|M.abscessus_ATCC_199 -----------------------------
MSMEG_0618|M.smegmatis_MC2_155 -----------------------------
Mflv_0326|M.gilvum_PYR-GCK -----------------------------
Mvan_0414|M.vanbaalenii_PYR-1 -----------------------------
Rv1806|M.tuberculosis_H37Rv -----------------------------
Mb1835|M.bovis_AF2122/97 -----------------------------
MMAR_4611|M.marinum_M -----------------------------
MUL_4462|M.ulcerans_Agy99 -----------------------------
MAV_1346|M.avium_104 SMATAGRGGYGLGAPRYGVKPTVMPKPAI
MLBr_01183|M.leprae_Br4923 -----------------------------