For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGGSSRRYPPELRERAVRMVAEIRGQHDSEWAAISEVARLLGVGCAETVRKWVRQAQVDAGARPGTTTEE SAELKRLRRDNAELRRANAILKTASAFFAAELDRPAR*
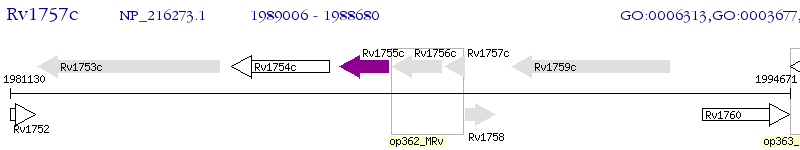
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1757c | - | - | 100% (108) | PUTATIVE TRANSPOSASE |
| M. tuberculosis H37Rv | Rv3474 | - | 3e-57 | 100.00% (108) | transposase IS6110 |
| M. tuberculosis H37Rv | Rv3381c | - | 3e-57 | 100.00% (108) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2839c | - | 3e-57 | 100.00% (108) | transposase |
| M. gilvum PYR-GCK | Mflv_0600 | - | 2e-21 | 49.50% (101) | Fis family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2087 | - | 1e-12 | 38.30% (94) | transposase |
| M. marinum M | MMAR_2547 | - | 1e-35 | 70.30% (101) | transposase, ISMyma03_aa1 |
| M. avium 104 | MAV_5035 | - | 1e-22 | 50.50% (101) | ISAfe7, transposase OrfA |
| M. smegmatis MC2 155 | MSMEG_2676 | - | 1e-36 | 69.23% (104) | IS1137, transposase orfA |
| M. thermoresistible (build 8) | TH_1992 | - | 2e-26 | 57.43% (101) | POSSIBLE TRANSPOSASE FOR INSERTION ELEMENT IS6110 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5972 | - | 2e-21 | 50.50% (101) | transposase IS3/IS911 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_5035|M.avium_104 -----------------------------------MPKEQSPGKPTTRRY
Mvan_5972|M.vanbaalenii_PYR-1 -----------------------------------MPKEQSVGKPTTRRY
Mflv_0600|M.gilvum_PYR-GCK -----------------------------------MPKEQSPGKPTTRRY
Rv1757c|M.tuberculosis_H37Rv -----------------------------------------MSGGSSRRY
Mb2839c|M.bovis_AF2122/97 -----------------------------------------MSGGSSRRY
MSMEG_2676|M.smegmatis_MC2_155 -----------------------------------------MGSKSSRRY
MMAR_2547|M.marinum_M -----------------------------------------MARAS--KY
TH_1992|M.thermoresistible__bu VLAKLIWQQHWGTWLKFGRADKLFTRLRAARLDHTVEAEIRRLAAVDXKY
MAB_2087|M.abscessus_ATCC_1997 ----------------------------MTSELACPKRAGWKDVSVPRPY
*
MAV_5035|M.avium_104 SAEEKAAAVRMVRTLRAEL-GTEQGTVQRVARQLGY-GVESVRTWVRQAD
Mvan_5972|M.vanbaalenii_PYR-1 SAEEKAAAVRMVRALRAEL-GIAQGTVQRVATQLGY-GVESVRTWVRQAD
Mflv_0600|M.gilvum_PYR-GCK SSEEKAAAVRMVRTLREEL-GTEQGTVTRVAHQLGY-GVESVRSWVRQAD
Rv1757c|M.tuberculosis_H37Rv PPELRERAVRMVAEIRGQH-DSEWAAISEVARLLGVGCAETVRKWVRQAQ
Mb2839c|M.bovis_AF2122/97 PPELRERAVRMVAEIRGQH-DSEWAAISEVARLLGVGCAETVRKWVRQAQ
MSMEG_2676|M.smegmatis_MC2_155 PDELKVRAVQMVADLRSET-VSEWEAMGRVADLLGVGTAETVRKWVRQAE
MMAR_2547|M.marinum_M PEELRERAVRMVAEIAPQY-GSQWAAICAVAGKLGVGTPETVRTWVRRAE
TH_1992|M.thermoresistible__bu SPEFRDRALRLLDTTMEDSGVSEFEAIKSVASKLGV-SQESVRRWRRKAE
MAB_2087|M.abscessus_ATCC_1997 PREFRDDVVRVARNRDDGV------TIEQIATDFGV-HPMTLTKWMRQAD
. * : .::: :: :* :* :: * *:*:
MAV_5035|M.avium_104 IDEGLAPGVTTAESKRVQELEQENRELKRANEILKRAASFFG-AELDRQH
Mvan_5972|M.vanbaalenii_PYR-1 IDEGVAPGVTTAESTRLKALEQENRELERANEILKRAASFFG-AELDRQH
Mflv_0600|M.gilvum_PYR-GCK IDDGHAPGVTSTESAKVKELEQEIRELKRANEILKRAASFFG-AELDRQH
Rv1757c|M.tuberculosis_H37Rv VDAGARPGTTTEESAELKRLRRDNAELRRANAILKTASAFFA-AELDRPA
Mb2839c|M.bovis_AF2122/97 VDAGARPGTTTEESAELKRLRRDNAELRRANAILKTASAFFA-AELDRPA
MSMEG_2676|M.smegmatis_MC2_155 IDAGSRAGQTSEESEVLRKLRRENAELKRANAILKAASVFFA-AELDRPS
MMAR_2547|M.marinum_M VDAGQRPGVSSEQSEELRRLKRENAELRRANEILKAAAIFFG-AELDRPA
TH_1992|M.thermoresistible__bu IDAGQRPGVTSTEHAEIRKLKRENAELRRANEILKSASAFFA-AELDRPA
MAB_2087|M.abscessus_ATCC_1997 VDEGAKPGKSTNDSADLRELRRRNRLLEQENEVLRRAAAYLSQANLPGKG
:* * .* :: : :: *.: *.: * :*: *: ::. *:*
MAV_5035|M.avium_104 KK--
Mvan_5972|M.vanbaalenii_PYR-1 KR--
Mflv_0600|M.gilvum_PYR-GCK KK--
Rv1757c|M.tuberculosis_H37Rv R---
Mb2839c|M.bovis_AF2122/97 R---
MSMEG_2676|M.smegmatis_MC2_155 Q---
MMAR_2547|M.marinum_M RR--
TH_1992|M.thermoresistible__bu TK--
MAB_2087|M.abscessus_ATCC_1997 STRS