For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVDDRQGRRGGRRPRSAAADNRPAFRDGPAIPPGIHARQLAPEIRRELSTLDRATADAVACHLVAAGELI DDDPEAALRHARAARVRASRIAAVREAVGIAAYRCGDWAQALAELRAARRMGSKSPLLALIADCERGLGR PQRAIELARGSEAVELSGDAADELRIVAAGARADLGQLEQALTVLSTPQLDPGRTGSTAARLFYAYAEIL LALGRGDEALQWFLRSAAADIDGVTDAEDRVDELGAREQK
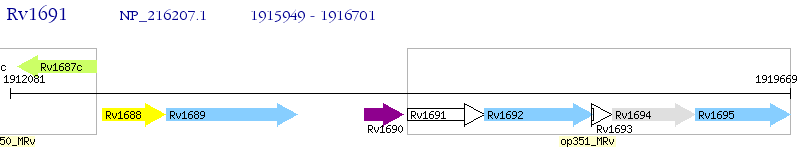
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1691 | - | - | 100% (250) | hypothetical protein Rv1691 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1717 | - | 1e-138 | 100.00% (250) | hypothetical protein Mb1717 |
| M. gilvum PYR-GCK | Mflv_3512 | - | 5e-95 | 66.79% (274) | hypothetical protein Mflv_3512 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2356 | - | 9e-91 | 70.20% (245) | hypothetical protein MAB_2356 |
| M. marinum M | MMAR_2496 | - | 1e-105 | 81.51% (238) | hypothetical protein MMAR_2496 |
| M. avium 104 | MAV_3080 | - | 1e-100 | 87.62% (210) | TPR repeat-containing protein |
| M. smegmatis MC2 155 | MSMEG_3754 | - | 1e-88 | 70.76% (236) | TPR repeat-containing protein |
| M. thermoresistible (build 8) | TH_1552 | - | 9e-84 | 79.19% (197) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1680 | - | 1e-105 | 81.09% (238) | hypothetical protein MUL_1680 |
| M. vanbaalenii PYR-1 | Mvan_3293 | - | 1e-95 | 77.16% (232) | hypothetical protein Mvan_3293 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1691|M.tuberculosis_H37Rv ------------------------------------------------MV
Mb1717|M.bovis_AF2122/97 ------------------------------------------------MV
MMAR_2496|M.marinum_M ------------------------------------------------MV
MUL_1680|M.ulcerans_Agy99 ------------------------------------------------MV
MAV_3080|M.avium_104 --------------------------------------------------
Mflv_3512|M.gilvum_PYR-GCK MRLFPR-----------AIHVMLLTNCINCES-------VDGGRILSFVP
Mvan_3293|M.vanbaalenii_PYR-1 ------------------------------MA-------PDLARILSVVP
TH_1552|M.thermoresistible__bu --------------------------------------------------
MSMEG_3754|M.smegmatis_MC2_155 MILFSAGPFSVSNSVRNPVRKMAEKACDYGWSGTGQYPFGDSAGRKAHVV
MAB_2356|M.abscessus_ATCC_1997 ---------------------MGQDASQRGPRQQGPRRQGPGRPARDAGA
Rv1691|M.tuberculosis_H37Rv DDRQGRRGGRRPRSAAADN-----------------------------RP
Mb1717|M.bovis_AF2122/97 DDRQGRRGGRRPRSAAADN-----------------------------RP
MMAR_2496|M.marinum_M DDRHGRGGQRRLRSASGDAPRRGPR----------RDRDGSWSGPGRARP
MUL_1680|M.ulcerans_Agy99 DDRHGRGGQRRLRSASGDAPRRGPR----------RDRDGSWSGPGRARP
MAV_3080|M.avium_104 --------------------------------------------------
Mflv_3512|M.gilvum_PYR-GCK HDNQGGRGERRPDRATT---GGSQR------PRRDNRSAPRTSGPNRARS
Mvan_3293|M.vanbaalenii_PYR-1 QDNQGGRGDRRPSRSSN---GNRER------PRRDQRSAPRSSGPNRARP
TH_1552|M.thermoresistible__bu --------------------------------------------------
MSMEG_3754|M.smegmatis_MC2_155 GDSQGGE-ERRPRRAASNRASNNQRGNRSFDPRSRDRRPPRSSGPGRARP
MAB_2356|M.abscessus_ATCC_1997 RRDDRGSGAARPRRSDERPQRSDER---------------GGDRPQRNQS
Rv1691|M.tuberculosis_H37Rv AFR------------DGPAIPPGIHARQLAPEIRRELSTLDRATADAVAC
Mb1717|M.bovis_AF2122/97 AFR------------DGPAIPPGIHARQLAPEIRRELSTLDRATADAVAC
MMAR_2496|M.marinum_M AQPQNASGDGTRARPEGPAVPPGVDAKQLAPEIRRELSTLDRATADAVAR
MUL_1680|M.ulcerans_Agy99 AQPQNASGDGTRARPEGPAVPPGVDAKQLAPEIRRELSTLDRATADAVAR
MAV_3080|M.avium_104 -----------------------MEAKQLAPDIRRELSTLDRATADAVAR
Mflv_3512|M.gilvum_PYR-GCK AQPRPDRVDPRF--ADAPPIPAEIEAKQLAPEIRGELTTLDRATADAVAR
Mvan_3293|M.vanbaalenii_PYR-1 AQPRTDGDESR---PSGPPIPAEIEAKQLAPEIRGELTTLDRSTADTVAR
TH_1552|M.thermoresistible__bu ------------------------------------LATLDRVTADAVAR
MSMEG_3754|M.smegmatis_MC2_155 AQP--ESAEDSQ--DSAPRLSADIEARQLAPEVRRELVTLDKATADYVAR
MAB_2356|M.abscessus_ATCC_1997 PRPRRTEDRPAP-RNFGPHIPDEVEAKQLAPDVRAELRTLDKNTAETVAR
* ***: **: **
Rv1691|M.tuberculosis_H37Rv HLVAAGELIDDDPEAALRHARAARVRASRIAAVREAVGIAAYRCGDWAQA
Mb1717|M.bovis_AF2122/97 HLVAAGELIDDDPEAALRHARAARVRASRIAAVREAVGIAAYRCGDWAQA
MMAR_2496|M.marinum_M HLVAAGELLDEDPQAALAHARAARARSSRIAVVREAVGIAAYHCGDWGQA
MUL_1680|M.ulcerans_Agy99 HLVAAGELLDEDPQAALAHARAARARSSRIAVVREAVGIAAYHCGDWGQA
MAV_3080|M.avium_104 HLVAAGELLDEDPEAALRHARAARARSSRIAAIREAVGIAAYHCGDWAQA
Mflv_3512|M.gilvum_PYR-GCK HLVAAGELLEDDPQAALAHAQAARARSGRIAAVREAVGIAAYQCGDWAQA
Mvan_3293|M.vanbaalenii_PYR-1 HLVAAGELLDDDPEAALAHARAARARSGRIAAVREAVGIAAYQCGDWAQA
TH_1552|M.thermoresistible__bu RLVAAGELLDEDPETALAHARAARARSGRIAAVREAVGIAAYHCGDWAQA
MSMEG_3754|M.smegmatis_MC2_155 HLVAAGNLLDEDPETALAHARAARGRAARIAAVREAVGIAAYHCGDWAQA
MAB_2356|M.abscessus_ATCC_1997 HLVTAGNLLDEDPQAALEHAKAAKSRAARIAAVREAVGIAAYHCGDWAQA
:**:**:*:::**::** **:**: *:.***.:*********:****.**
Rv1691|M.tuberculosis_H37Rv LAELRAARRMGSKSPLLALIADCERGLGRPQRAIELARGSEAVELSGDAA
Mb1717|M.bovis_AF2122/97 LAELRAARRMGSKSPLLALIADCERGLGRPQRAIELARGSEAVELSGDAA
MMAR_2496|M.marinum_M LAELRAARRMGSKSALLALMADCERGLGRPQRAIDLARGPEATQLSGDDA
MUL_1680|M.ulcerans_Agy99 LAELRAARRMGSKSALLALMADCERGLGRPQRAIDLARGPEATQLSGDDA
MAV_3080|M.avium_104 LAELRAARRMGSKSALLALIADCERGLGRPERAIELARGPEAAELSGDDA
Mflv_3512|M.gilvum_PYR-GCK LAELRAARRMGSRSPLLALIADCERGVGRPERAIELARGEEAAALTGEDA
Mvan_3293|M.vanbaalenii_PYR-1 LAELRAARRMGSRSPLLALIADCERGVGRPERAIELARSPEAAELTGDDA
TH_1552|M.thermoresistible__bu LSEFRAARRMGSKSPLLPLIADCERGVGRPERAIELARSPEAAELTGDDA
MSMEG_3754|M.smegmatis_MC2_155 LAELRAARRMGSKSPLLPMIADCERGVGRPERAVELARSPEAEALTGEDA
MAB_2356|M.abscessus_ATCC_1997 LSELRAARRMGSKSPLLALIADCERGVGRPERAIELARGDEAKALTGDAA
*:*:********:*.**.::******:***:**::***. ** *:*: *
Rv1691|M.tuberculosis_H37Rv DELRIVAAGARADLGQLEQALTVLSTPQLDPGRTGSTAARLFYAYAEILL
Mb1717|M.bovis_AF2122/97 DELRIVAAGARADLGQLEQALTVLSTPQLDPGRTGSTAARLFYAYAEILL
MMAR_2496|M.marinum_M DELRIVAAGARADLGQLEQALTLLSTPQLDPDRTGSTAARLFYAYADTLL
MUL_1680|M.ulcerans_Agy99 DELRIVAAGALADLGQLEQALTLLSTPQLDPDRTGSTAARLFYAYADTLL
MAV_3080|M.avium_104 DELRIVAAGARADLGQLEQALTILSTPQLDSTRTGSTAARLFYAYADTLL
Mflv_3512|M.gilvum_PYR-GCK DELRIVVAGARSDLGQHEQALAILSTPTLDPSKSGQTAARLFYAYADTLV
Mvan_3293|M.vanbaalenii_PYR-1 DELRMVVAGARSDLGQHEQALAVLSSPALDPTRTGPTAARLFYAYAETLL
TH_1552|M.thermoresistible__bu DELRIVVAGARSDLGQHEQALAVLASPPLDPKRTGTTAARLFYAYADTLL
MSMEG_3754|M.smegmatis_MC2_155 DELRIVVAGARADLGQFEQALTLLSTPPLDPTSRGQTAARLFYVYADLLL
MAB_2356|M.abscessus_ATCC_1997 DELRIVVAGARADMGQLDQALAVLSTPAPDPSRVGEIAARLFYAYADILL
****:*.*** :*:** :***::*::* *. * ******.**: *:
Rv1691|M.tuberculosis_H37Rv ALGRGDEALQWFLRSAAADIDGVTDAEDRVDELGAREQK
Mb1717|M.bovis_AF2122/97 ALGRGDEALQWFLRSAAADIDGVTDAEDRVDELGAREQK
MMAR_2496|M.marinum_M ALGRRDEALHWFLRSAAADSEGVTDAEDRVDELAE----
MUL_1680|M.ulcerans_Agy99 ALGRRDEALHWFLRSAAADSEGVTDAEDRVDELAE----
MAV_3080|M.avium_104 ALGRNDEALQWFLHAAAADLDGVTDAEDRVSELA-----
Mflv_3512|M.gilvum_PYR-GCK ALDRRDEALQWFLHAAKADVDGITDAEERITELS-----
Mvan_3293|M.vanbaalenii_PYR-1 ALGREAEALQWFLHAAAADVEGVTDAEERVTELS-----
TH_1552|M.thermoresistible__bu KLGRRDEALQWFLHAAAADVAGATDAEDRVTELS-----
MSMEG_3754|M.smegmatis_MC2_155 AVERTDEALQWFIHAAEADVEGVTDAEERITELS-----
MAB_2356|M.abscessus_ATCC_1997 ALDRKAEAVQWFLHAADADVEGHTDAEDRAAELA-----
: * **::**:::* ** * ****:* **.