For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVLNRTDTLVDELTADITNTPLGYGGVDGDERWAAEIRRLAHLRGATVLAHNYQLPAIQDVADHVGDSL ALSRVAAEAPEDTIVFCGVHFMAETAKILSPHKTVLIPDQRAGCSLADSITPDELRAWKDEHPGAVVVSY VNTTAAVKALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRVTGRKNLHVWAGECHVHAGINGD ELADQARAHPDAELFVHPECGCATSALYLAGEGAFPAERVKILSTGGMLEAAHTTRARQVLVATEVGMLH QLRRAAPEVDFRAVNDRASCKYMKMITPAALLRCLVEGADEVHVDPGIAASGRRSVQRMIEIGHPGGGE
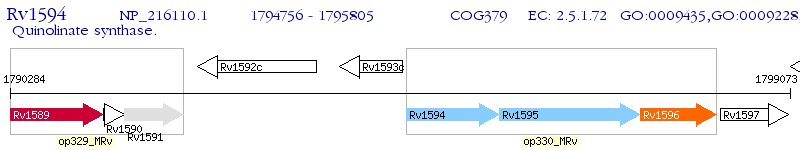
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1594 | nadA | - | 100% (349) | quinolinate synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1620 | nadA | 0.0 | 100.00% (349) | quinolinate synthetase |
| M. gilvum PYR-GCK | Mflv_3619 | - | 1e-169 | 84.86% (350) | quinolinate synthetase |
| M. leprae Br4923 | MLBr_01225 | nadA | 1e-174 | 84.38% (352) | quinolinate synthetase |
| M. abscessus ATCC 19977 | MAB_2674c | - | 1e-166 | 84.16% (341) | quinolinate synthetase |
| M. marinum M | MMAR_2391 | nadA | 1e-180 | 88.07% (352) | quinolinate synthetase NadA |
| M. avium 104 | MAV_3191 | nadA | 0.0 | 89.40% (349) | quinolinate synthetase |
| M. smegmatis MC2 155 | MSMEG_3199 | nadA | 1e-172 | 85.67% (349) | quinolinate synthetase |
| M. thermoresistible (build 8) | TH_0809 | nadA | 1e-161 | 85.71% (322) | Probable quinolinate synthetase nadA |
| M. ulcerans Agy99 | MUL_1568 | nadA | 1e-180 | 87.78% (352) | quinolinate synthetase |
| M. vanbaalenii PYR-1 | Mvan_2798 | - | 1e-174 | 88.72% (337) | quinolinate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3619|M.gilvum_PYR-GCK MTVLAGTF-----DAALADRVVDGPGGFSGVDGDAAWADEVRRLLKLRGA
Mvan_2798|M.vanbaalenii_PYR-1 MTVLAGAF-----DTELTAGIVDGPGGFTGVEGDEKWAAEVRRLLKLRGA
MSMEG_3199|M.smegmatis_MC2_155 MTVLNGTP-----AGALADRIVDGPGGYSGIDGDEQWAAEVRRLVELRGA
MAB_2674c|M.abscessus_ATCC_199 MTAAD----------VVLDGVIDGPGGYTGVEGDARWAEMVRRLAEQRGA
Rv1594|M.tuberculosis_H37Rv MTVLNRTDTLVDE---LTADITNTPLGYGGVDGDERWAAEIRRLAHLRGA
Mb1620|M.bovis_AF2122/97 MTVLNRTDTLVDE---LTADITNTPLGYGGVDGDERWAAEIRRLAHLRGA
MMAR_2391|M.marinum_M MTVLNRMDTLACDVSGMTDRISDSPLGYVGVDGDQQWAAEVRRLARLRGA
MUL_1568|M.ulcerans_Agy99 MTVLNRMDTLACDVSGMTDRISDSPLGYVGVDGDQQWAAEVRRLARLRGA
MAV_3191|M.avium_104 MTVLNRMDTLAED---MVADITDSPTGYTGVDGDAQWAAEVRRLAKLRGA
MLBr_01225|M.leprae_Br4923 MTVLNGMEPLAGDMTVVIAGITDSPVGYAGVDGDEQWATEIRRLTRLRGA
TH_0809|M.thermoresistible__bu MTVLDQRLS--------HDDASDWEASAPDVEPTAEWAAEIRRLADARGA
**. : . .:: ** :*** ***
Mflv_3619|M.gilvum_PYR-GCK TLLAHNYQLPEIQDVADHVGDSLALSRIAAEVPEDTIVFAGVHFMAETAK
Mvan_2798|M.vanbaalenii_PYR-1 TLLAHNYQLPAIQDVADHVGDSLALSRIAAEAPEDTIVFAGVHFMAETAK
MSMEG_3199|M.smegmatis_MC2_155 TLLAHNYQLPAIQDVADHVGDSLALSRIAAEAPEDTIVFCGVHFMAETAK
MAB_2674c|M.abscessus_ATCC_199 TVLAHNYQLPAIQDVADHVGDSLALSRIAAEAPEDTIVFCGVHFMAETAK
Rv1594|M.tuberculosis_H37Rv TVLAHNYQLPAIQDVADHVGDSLALSRVAAEAPEDTIVFCGVHFMAETAK
Mb1620|M.bovis_AF2122/97 TVLAHNYQLPAIQDVADHVGDSLALSRVAAEAPEDTIVFCGVHFMAETAK
MMAR_2391|M.marinum_M TLLAHNYQLPAIQDVADHVGDSLALSRIAAEVPEGTIVFCGVHFMAETAK
MUL_1568|M.ulcerans_Agy99 TLLAHNYQLPAIQDVADHVGDSLALSRIAAEVPEGTIVFCGVHFMAETAK
MAV_3191|M.avium_104 TILAHNYQLPEIQDVADHVGDSLALSRIAAEAPEDTIVFCGVHFMAETAK
MLBr_01225|M.leprae_Br4923 TVLAHNYQLPAIQDIADYVGDSLALSRIAAEVPEETIVFCGVHFMAETAK
TH_0809|M.thermoresistible__bu TLLAHNYQLPAIQDVADHVGDSLALSRIAAEAPEGTIVFCGVHFMAETAK
*:******** ***:**:*********:***.** ****.**********
Mflv_3619|M.gilvum_PYR-GCK ILSPEKTVLIPDQRAGCSLADSITADELRAWKAEHPGAVVVSYVNTTAAV
Mvan_2798|M.vanbaalenii_PYR-1 ILSPDKTVLIPDQRAGCSLADSITADELRAWKAEHPGAVVVSYVNTTAAV
MSMEG_3199|M.smegmatis_MC2_155 ILSPDKTVLIPDQRAGCSLADSITAEELQAWKDDHPGAVVVSYVNTTAAV
MAB_2674c|M.abscessus_ATCC_199 ILSPEKTVLIPDQRAGCSLADSITADQLRDWKAEFPDAVVVSYVNTTAAV
Rv1594|M.tuberculosis_H37Rv ILSPHKTVLIPDQRAGCSLADSITPDELRAWKDEHPGAVVVSYVNTTAAV
Mb1620|M.bovis_AF2122/97 ILSPHKTVLIPDQRAGCSLADSITPDELRAWKDEHPGAVVVSYVNTTAAV
MMAR_2391|M.marinum_M ILSPNKTVLIPDQRAGCSLADSISPDELRAWKAEHPGAVVVSYVNTTAEV
MUL_1568|M.ulcerans_Agy99 ILSPNKTVLIPDQRAGCSLADSISPDELRAWKAEHPGAVVVSYVNTTAEV
MAV_3191|M.avium_104 ILSPDKTVLIPDQRAGCSLADSITPEDLRAWKDEHPGAVVVSYVNTTAAV
MLBr_01225|M.leprae_Br4923 ILSPNKTVLIPDQRAGCSLADSITPDELCAWKDEHPGAAVVSYVNTTAEV
TH_0809|M.thermoresistible__bu ILSPDKTVLIPDARAGCSLADSITAEDLRAWKAEHPGAVVVSYVNTTAAV
****.******* **********:.::* ** :.*.*.********* *
Mflv_3619|M.gilvum_PYR-GCK KAETDICCTSSNAVEVVASIPADREVLFCPDQFLGAHVRRITGRTNLHVW
Mvan_2798|M.vanbaalenii_PYR-1 KAETDICCTSSNAVEVVASIPEDREVLFCPDQFLGAHVRRVTGRKNLHVW
MSMEG_3199|M.smegmatis_MC2_155 KALTDICCTSSNAVEVVASIPEDREVLFCPDQFLGAHVRRVTGRKNMHVW
MAB_2674c|M.abscessus_ATCC_199 KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVKRVTGRQNLHIW
Rv1594|M.tuberculosis_H37Rv KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRVTGRKNLHVW
Mb1620|M.bovis_AF2122/97 KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRVTGRKNLHVW
MMAR_2391|M.marinum_M KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRETGRENLHVW
MUL_1568|M.ulcerans_Agy99 KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRSTGRENLHVW
MAV_3191|M.avium_104 KALTDICCTSSNAVDVVASIDPDREVLFCPDQFLGAHVRRMTGRENLHVW
MLBr_01225|M.leprae_Br4923 KALTDICCTSSNAVDVVESIDPSREVLFCPDQFLGAHVRRVTGRKNVYVW
TH_0809|M.thermoresistible__bu KAETDICCTSSNAVDVVRSIPADREVLFCPDQFLGAHVRRVTGRENMHVW
** ***********:** ** .***************:* *** *:::*
Mflv_3619|M.gilvum_PYR-GCK AGECHVHAGINGDELADQARAHPDAELYVHPECGCATSALYLAGEGAFPE
Mvan_2798|M.vanbaalenii_PYR-1 AGECHVHAGINGDELADQARAHPDAELFVHPECGCATSALYLAGEGAFPE
MSMEG_3199|M.smegmatis_MC2_155 AGECHVHAGINGDELAAQARSHPDAELFVHPECGCATSALYLAGEGAVPE
MAB_2674c|M.abscessus_ATCC_199 AGECHVHAGINGDELAGQARSHPNAELFVHPECGCATSALYLAGEGAVPD
Rv1594|M.tuberculosis_H37Rv AGECHVHAGINGDELADQARAHPDAELFVHPECGCATSALYLAGEGAFPA
Mb1620|M.bovis_AF2122/97 AGECHVHAGINGDELADQARAHPDAELFVHPECGCATSALYLAGEGAFPA
MMAR_2391|M.marinum_M AGECHVHAGINGDELSEQARANPDAELFVHPECGCATSALYLAGEGAVPA
MUL_1568|M.ulcerans_Agy99 AGECHVHVGINGDELSEQARANPDAELFVHPECGCATSALYLAGEGAVPA
MAV_3191|M.avium_104 AGECHVHAGINGDELGERARANPDAELFVHPECGCATSALYLAGEGAFPA
MLBr_01225|M.leprae_Br4923 MGECHVHAGINGDELVDQARANPDAELFVHPECGCSTSALYLAGEGAFPP
TH_0809|M.thermoresistible__bu AGECHVHAGINGEELSDQARSHPDAELFVHPECGCATSALYLAGEGAFPA
******.****:** :**::*:***:*******:***********.*
Mflv_3619|M.gilvum_PYR-GCK DRVKILSTGGMLDAARETGARQVLVATEVGMLHQLRRAAPDVDFRAVNDR
Mvan_2798|M.vanbaalenii_PYR-1 DRVKILSTGGMLDAARETGARQVLVATEVGMLHQLRRAAPEIDFRAVNDR
MSMEG_3199|M.smegmatis_MC2_155 DRVKILSTGGMLDAARETSARQVLVATEIGMLHQLRRAAPEVDFLAVNDR
MAB_2674c|M.abscessus_ATCC_199 DRVKILSTGGMLDAARETRAREVLVATEVGMLHQLRKAAPDVNFLAVNDR
Rv1594|M.tuberculosis_H37Rv ERVKILSTGGMLEAAHTTRARQVLVATEVGMLHQLRRAAPEVDFRAVNDR
Mb1620|M.bovis_AF2122/97 ERVKILSTGGMLEAAHTTRARQVLVATEVGMLHQLRRAAPEVDFRAVNDR
MMAR_2391|M.marinum_M ERVKILSTGGMLDAAHETRARKVLVATEVGMLHQLRRAAPQVDFQAVNDR
MUL_1568|M.ulcerans_Agy99 ERVKILSTGGMLDAAHETRARKVLVATEVGMLHQLRRAAPQVDFQAVNDR
MAV_3191|M.avium_104 DRVKILSTGGMLDAANTTSARKVLVATEVGMLYQLRRAAPQVDFQAVNDR
MLBr_01225|M.leprae_Br4923 DRVKILSTGGMLTAARQTQYRKILVATEVGMLYQLRRAAPEIDFRAVNDR
TH_0809|M.thermoresistible__bu ERVKILSTGGMIDAARQSQARQVLVATEVGMLHQLRRAAPEIDFRAVNDR
:**********: **. : *::*****:***:***:***:::* *****
Mflv_3619|M.gilvum_PYR-GCK ASCRYMKMITPAAMLRCLVEGADEVHVDPDMAAKARLSVQRMIEIGQPGG
Mvan_2798|M.vanbaalenii_PYR-1 ASCRFMKMITPAAMLRCLIEGADEVHVDPDVAASARLSVQRMIEIGQPGG
MSMEG_3199|M.smegmatis_MC2_155 ASCTYMKMITPAALLRCLVEGADEVHVDPETARLGRASVQRMIEIGQPGG
MAB_2674c|M.abscessus_ATCC_199 ASCTYMKMITPAALLRCLVEGRDEVDVDPDTARLARRSVQRMIEIGQPGG
Rv1594|M.tuberculosis_H37Rv ASCKYMKMITPAALLRCLVEGADEVHVDPGIAASGRRSVQRMIEIGHPGG
Mb1620|M.bovis_AF2122/97 ASCKYMKMITPAALLRCLVEGADEVHVDPGIAASGRRSVQRMIEIGHPGG
MMAR_2391|M.marinum_M ASCKYMKMITPAALLRCLVEGADEVHVDPDVAAAGRLSVQRMIEIGQPGG
MUL_1568|M.ulcerans_Agy99 ASCKYMKMITPAALLRCLVEGADEVHVDPDVAAAGRLSVQRMIEIGQPGG
MAV_3191|M.avium_104 ASCRYMKMITPAALLRCLVEGADEVHVDPGIAAAGRRSVQRMIEIGQPGG
MLBr_01225|M.leprae_Br4923 ASCKYMKMITPGALLRCLVEGTDEVHVDSEIAAAGRRSVQRMIEIGLPGG
TH_0809|M.thermoresistible__bu ASCRFMKMITPGALLRCLQEGRDVIEVDPEIAEKARRSVQRMIEIGQPGS
*** :******.*:**** ** * :.**. * .* ********* **.
Mflv_3619|M.gilvum_PYR-GCK GE
Mvan_2798|M.vanbaalenii_PYR-1 GE
MSMEG_3199|M.smegmatis_MC2_155 GE
MAB_2674c|M.abscessus_ATCC_199 GE
Rv1594|M.tuberculosis_H37Rv GE
Mb1620|M.bovis_AF2122/97 GE
MMAR_2391|M.marinum_M GE
MUL_1568|M.ulcerans_Agy99 GE
MAV_3191|M.avium_104 GE
MLBr_01225|M.leprae_Br4923 GE
TH_0809|M.thermoresistible__bu GE
**