For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVDPTATDSPKVSIVSISYNQEEYIREALDGFAAQRTEFPVEVIIADDASTDATPRIIGEYAARYPQLFR PILRQTNIGVHANFKDVLSAARGEYLALCEGDDYWTDPLKLSKQVKYLDRHPETTVCFHPVRVIYEDGAK DSEFPPLSWRRDLSVDALLARNFIQTNSVVYRRQPSYDDIPANVMPIDWYLHVRHAVGGEIAMLPETMAV YRRHAHGIWHSAYTDRRKFWETRGHGMAATLEAMLDLVHGHREREAIVGEVSAWVLREIGKTPGRQGRAL LLKSIADHPRMTMLSLQHRWAQTPWRRFKRRLSTELSSLAALAYATRRRALEGRDGGYRETTSPPTGRGR NVRGSHA
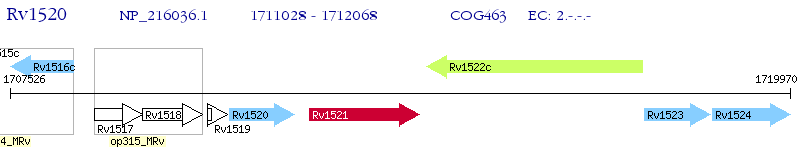
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1520 | - | - | 100% (346) | probable sugar transferase |
| M. tuberculosis H37Rv | Rv1516c | - | e-113 | 63.09% (317) | sugar transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1547 | - | 0.0 | 100.00% (346) | sugar transferase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3255 | - | 2e-98 | 53.87% (336) | glycosyltransferase GtfTB |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3097 | - | 7e-06 | 25.84% (209) | PUTATIVE transferase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1520|M.tuberculosis_H37Rv ---MSIVSISYNQEEYIREALDGFAAQRTEFPVEVIIADDASTDATPRII
Mb1547|M.bovis_AF2122/97 ---MSIVSISYNQEEYIREALDGFAAQRTEFPVEVIIADDASTDATPRII
MAV_3255|M.avium_104 ------MSTTHNQQAYVRDTLDGFLAQRVDFPMEVIVADDASTDATPRII
TH_3097|M.thermoresistible__bu LTASTRLSFVIASRNRAAELASVVNRLLDTTPCPVIVVDNASDDHSVDVI
:* .. : . . * **:.*:** * : :*
Rv1520|M.tuberculosis_H37Rv GEYAARYPQLFRPILRQTNIGVHANFKDVLSAARGEYLALCEGDDYWTDP
Mb1547|M.bovis_AF2122/97 GEYAARYPQLFRPILRQTNIGVHANFKDVLSAARGEYLALCEGDDYWTDP
MAV_3255|M.avium_104 QDYADRHPNLFRPILRSRNVGLNANLTGALSAARGEYIALCEGDDYWVDP
TH_3097|M.thermoresistible__bu TRIAARSANRVQVIELDSNRGAVG-RNAGVAASRTPYVAFCD-DDSWWEP
* * .: .: * . * * . . ::*:* *:*:*: ** * :*
Rv1520|M.tuberculosis_H37Rv LKLSKQVKYLDRHPETTVCFHPVRVIYEDGAKDSE---------------
Mb1547|M.bovis_AF2122/97 LKLSKQVKYLDRHPETTVCFHPVRVIYEDGAKDSE---------------
MAV_3255|M.avium_104 LKLTKQVALLDEDPDTTVCFHPVQVVWTHECAEDEKLVHTLYRKFEEIFL
TH_3097|M.thermoresistible__bu DALALGVETFDRYP--RVGLLAART-------------------------
*: * :*. * * : ..:.
Rv1520|M.tuberculosis_H37Rv --FPPLSWRRDLSVDALLARNFIQTNSVVYRRQPSYDDIPANVMPIDWYL
Mb1547|M.bovis_AF2122/97 --FPPLSWRRDLSVDALLARNFIQTNSVVYRRQPSYDDIPANVMPIDWYL
MAV_3255|M.avium_104 PKFPPPFRAGDLSFETLISRNFIQTNSVMYRRLPRYDDIPADVMPLDWYL
TH_3097|M.thermoresistible__bu ---------------TVLPGGRIDPFSIELARSP----------------
:::. . *:. *: * *
Rv1520|M.tuberculosis_H37Rv HVRHAVGGEIAMLPETMAVYRRHAHGIWHSAYTDRRKFWETRGHGMAATL
Mb1547|M.bovis_AF2122/97 HVRHAVGGEIAMLPETMAVYRRHAHGIWHSAYTDRRKFWETRGHGMAATL
MAV_3255|M.avium_104 HVRHAAEGAIAMLPETMAVYRRHPRGMWYKSITDPATFWRMQGPGHAATL
TH_3097|M.thermoresistible__bu -----------------LGHRPDLPGPSILGFMSCAAMVRTAAFQAAGGF
:* . * . . : . . *. :
Rv1520|M.tuberculosis_H37Rv EAMLDLVHGHREREAIVGEVSAWVLREIG-KTPGRQGRALLLKSIADHPR
Mb1547|M.bovis_AF2122/97 EAMLDLVHGHREREAIVGEVSAWVLREIG-KTPGRQGRALLLKSIADHPR
MAV_3255|M.avium_104 DAMLDVVSGDPVHESIVAKNADWVLGAIAKQVPDPEGQQVLTQTTAKYPR
TH_3097|M.thermoresistible__bu SDILHFR-GEEQLLALDMAALGWELCYLAELLAVHQPSSVRATTXXXAPR
. :*.. *. :: * * :. . : : : **
Rv1520|M.tuberculosis_H37Rv MTMLSLQHRWAQTPWRRFKRRLSTELSSLAALAYATRRRALEGRDGGYRE
Mb1547|M.bovis_AF2122/97 MTMLSLQHRWAQTPWRRFKRRLSTELSSLAALAYATRRRALEGRDGGYRE
MAV_3255|M.avium_104 IAALALRNRCTKTLGGRVKR-LRRKLS---------------------RN
TH_3097|M.thermoresistible__bu SLWWPKRSPWRRCRRRRIR-------------------------------
. : : *.:
Rv1520|M.tuberculosis_H37Rv TTSPPTGRGRNVRGSHA-
Mb1547|M.bovis_AF2122/97 TTSPPTGRGRNVRGSHA-
MAV_3255|M.avium_104 ATAPNSDAERRIDEPLSA
TH_3097|M.thermoresistible__bu ------------------