For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQGAVAGLVFLAVLVIFAIIVVAKSVALIPQAEAAVIERLGRYSRTVSGQLTLLVPFIDRVRARVDLRER VVSFPPQPVITEDNLTLNIDTVVYFQVTVPQAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQ INAQLRGVLDEATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTREAAIKQAEGQK QAQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLQAQGQAKAIEKTFAAIKAGRPTPEMLAYQYLQ TLPEMARGDANKVWVVPSDFNAALQGFTRLLGKPGEDGVFRFEPSPVEDQPKHAADGDDAEVAGWFSTDT DPSIARAVATAEAIARKPVEGSLGTPPRLTQ
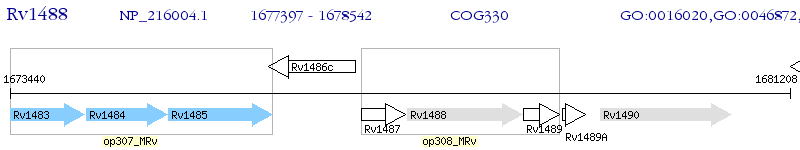
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1488 | - | - | 100% (381) | POSSIBLE EXPORTED CONSERVED PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1524 | - | 0.0 | 100.00% (381) | hypothetical protein Mb1524 |
| M. gilvum PYR-GCK | Mflv_3653 | - | 1e-179 | 84.64% (384) | band 7 protein |
| M. leprae Br4923 | MLBr_01802 | - | 0.0 | 89.54% (373) | hypothetical protein MLBr_01802 |
| M. abscessus ATCC 19977 | MAB_2718c | - | 1e-166 | 80.00% (365) | hypothetical protein MAB_2718c |
| M. marinum M | MMAR_2294 | - | 0.0 | 94.15% (376) | hypothetical protein MMAR_2294 |
| M. avium 104 | MAV_3290 | - | 0.0 | 92.51% (374) | secreted protein |
| M. smegmatis MC2 155 | MSMEG_3155 | - | 1e-178 | 85.53% (380) | band 7 protein |
| M. thermoresistible (build 8) | TH_1418 | - | 1e-167 | 79.84% (377) | POSSIBLE EXPORTED CONSERVED PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2759 | - | 0.0 | 86.72% (384) | band 7 protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3653|M.gilvum_PYR-GCK MDGALAGLVLLAVLVVFAAVIVAKSVALIPQAEAAVIERLGRYSKTVSGQ
Mvan_2759|M.vanbaalenii_PYR-1 MEGAVAGLILLAVLVVFAVIVVAKSVALIPQAEAAVIERLGRYSKTVSGQ
Rv1488|M.tuberculosis_H37Rv MQGAVAGLVFLAVLVIFAIIVVAKSVALIPQAEAAVIERLGRYSRTVSGQ
Mb1524|M.bovis_AF2122/97 MQGAVAGLVFLAVLVIFAIIVVAKSVALIPQAEAAVIERLGRYSRTVSGQ
MMAR_2294|M.marinum_M MDDAVVGLVFLAVLVIFAIIVVAKSVALIPQAEAAVIERLGRYSRTVSGQ
MAV_3290|M.avium_104 MQGAVAGLVLLAVLVIFAIVVVAKSVALIPQAEAAVIERLGRYSRTVSGQ
MLBr_01802|M.leprae_Br4923 MQGVVAGLVLLAVLTIFAIVVVAKSIVLIPQAEAAVVERLGRYGRTVSGQ
MSMEG_3155|M.smegmatis_MC2_155 MEGAVAGLVLLVVLVVFAIIVVAKSVALIPQAEAAVIERLGRYSKTVSGQ
TH_1418|M.thermoresistible__bu MDGAVAGLVILIVLVVFAAILVAKSVALIPQAEAAVIERLGRYSRTVSGQ
MAB_2718c|M.abscessus_ATCC_199 -MGVAAGVFVLIVLIILGVTIVLKSVALVPQAEAAVIERLGRYSKTVSGQ
.. .*:..* ** ::. :* **:.*:*******:******.:*****
Mflv_3653|M.gilvum_PYR-GCK LTLLLPFVDKIRARVDLRERVVSFPPQPVITEDNLTVNIDTVVYFQVTNP
Mvan_2759|M.vanbaalenii_PYR-1 LTLLLPFIDRIRARVDLRERVVSFPPQPVITEDNLTVNIDTVVYFQVTNP
Rv1488|M.tuberculosis_H37Rv LTLLVPFIDRVRARVDLRERVVSFPPQPVITEDNLTLNIDTVVYFQVTVP
Mb1524|M.bovis_AF2122/97 LTLLVPFIDRVRARVDLRERVVSFPPQPVITEDNLTLNIDTVVYFQVTVP
MMAR_2294|M.marinum_M LTLLVPFIDRVRARVDLRERVVSFPPQPVITEDNLTLNIDTVVYFQVTVP
MAV_3290|M.avium_104 LTLLVPFIDRIRARVDLRERVVSFPPQPVITEDNLTLNIDTVVYFQVTVP
MLBr_01802|M.leprae_Br4923 LTLLVLFIDRVRARVDLRERVVSFPPQPVITEDNLTLNIDTVVYFQVTVP
MSMEG_3155|M.smegmatis_MC2_155 LTLLVPFIDRIRARVDLRERVVSFPPQPVITEDNLTVQIDTVVYFQVTNP
TH_1418|M.thermoresistible__bu LTLLVPFIDRIRARVDLRERVVSFPPRAVITQDNLTVSIDTVVYFRVNNP
MAB_2718c|M.abscessus_ATCC_199 LTILVPFVDRIRAKVDLRERVVSFPPQPVITEDNLTVNIDTVVYFQVTNP
**:*: *:*::**:************:.***:****:.*******:*. *
Mflv_3653|M.gilvum_PYR-GCK QAAVYQISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDSINGQLRGVLD
Mvan_2759|M.vanbaalenii_PYR-1 QAAVYQISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINGQLRGVLD
Rv1488|M.tuberculosis_H37Rv QAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQLRGVLD
Mb1524|M.bovis_AF2122/97 QAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQLRGVLD
MMAR_2294|M.marinum_M QAAVYEISNYIVGVEQLATTTLRNVVGGMTLEQTLTSRDQINGQLRGVLD
MAV_3290|M.avium_104 QAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINGQLRGVLD
MLBr_01802|M.leprae_Br4923 QAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQLRGVLD
MSMEG_3155|M.smegmatis_MC2_155 QAAVYQISNYIVGVEQLATTTLRNLVGGMTLEQTLTSRDQINTALRGVLD
TH_1418|M.thermoresistible__bu HDAVYQISNYIAAVEELTTTTLRNVVGGMTLEEALTSRDRINEQLRAVLD
MAB_2718c|M.abscessus_ATCC_199 QAAVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINGQLRGVLD
: ***:*****..**:*:******:*******::***** ** **.***
Mflv_3653|M.gilvum_PYR-GCK EATNRWGLRVARVELRSIDPPPSIQDSMEKQMRADREKRAMILTAEGSRE
Mvan_2759|M.vanbaalenii_PYR-1 EATGRWGLRVARVELRSIDPPPSIQDSMEKQMRADREKRAMILTAEGSRE
Rv1488|M.tuberculosis_H37Rv EATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTRE
Mb1524|M.bovis_AF2122/97 EATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTRE
MMAR_2294|M.marinum_M EATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTRE
MAV_3290|M.avium_104 EATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGMRE
MLBr_01802|M.leprae_Br4923 EATGRWGLRVARVELRSIDPPPSIQTSMEKQMKADREKRAMILTAEGTRE
MSMEG_3155|M.smegmatis_MC2_155 EATGRWGLRVARVELRSIDPPPSIQDSMEKQMRADREKRAMILTAEGSRE
TH_1418|M.thermoresistible__bu GATDRWGLRVQRVELKAIDPPPTIQDAMEKQMRADREKRAMILTAEGQRE
MAB_2718c|M.abscessus_ATCC_199 EATGRWGLRVARVELRSIDPPPSVQESMEKQMKADREKRAMILNAEGVRE
**.****** ****::*****::* :*****:**********.*** **
Mflv_3653|M.gilvum_PYR-GCK AAIKQAEGQKQAQILAAEGAKQASILAAEGDRQSRMLRAQGERAAAYLQA
Mvan_2759|M.vanbaalenii_PYR-1 AAIKQAEGQKQAQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLQA
Rv1488|M.tuberculosis_H37Rv AAIKQAEGQKQAQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLQA
Mb1524|M.bovis_AF2122/97 AAIKQAEGQKQAQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLQA
MMAR_2294|M.marinum_M AAIKQAEGQKQSQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLRA
MAV_3290|M.avium_104 SAIKEAEGQKQAQILAAEGAKQAAILAAEADRQSRMLRAQGERAAAYLQA
MLBr_01802|M.leprae_Br4923 AAIKQAEGNKQAQILAAEGAKQAVILAAEADRQSRMLRAQGKRAAAYLQA
MSMEG_3155|M.smegmatis_MC2_155 AAIKQAEGQKQAQILAAEGAKQAAILTAEADRQSRMLRAQGERAAAYLQA
TH_1418|M.thermoresistible__bu SAIKEAEGQKQAQILAAEGAKQAAILTAEAERQSRILRAQGERAAQYLQA
MAB_2718c|M.abscessus_ATCC_199 ASIKQAEGAKQSQILAAEGAKQAAILSAEADRQSRILRAEGERAAQYLQA
::**:*** **:*********** **:**.:****:***:*:*** **:*
Mflv_3653|M.gilvum_PYR-GCK QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQTLPQMAKGEANKVWLVPSDF
Mvan_2759|M.vanbaalenii_PYR-1 QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQTLPQMAKGEANKVWLVPSDF
Rv1488|M.tuberculosis_H37Rv QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQTLPEMARGDANKVWVVPSDF
Mb1524|M.bovis_AF2122/97 QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQTLPEMARGDANKVWVVPSDF
MMAR_2294|M.marinum_M QGEAKAIQKTFAAIKAGRPTPEMLAYQYLQTLPEMARGDANKVWVVPSDF
MAV_3290|M.avium_104 QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQTLPEMARGDANKVWVVPSDF
MLBr_01802|M.leprae_Br4923 QGQAKAIEKTFAAIKAGRPTPEMLAYQYLQILPQMARGDANKVWVVPSDF
MSMEG_3155|M.smegmatis_MC2_155 QGQAKAIEKTFAAIKAGRPTPELLAYQYLQTLPQMAKGEANKVWLVPSDF
TH_1418|M.thermoresistible__bu QGQAKAIEKTFAAIKAGRPTPELLAYQYLQTLPKIAEGEASKVWIVPNDF
MAB_2718c|M.abscessus_ATCC_199 QGQAKAIEKVFAAVKSGKPTPELLAYQYLQTLPKMAEGEANKVWLIPSDF
**:****:*.***:*:*:****:******* **::*.*:*.***::*.**
Mflv_3653|M.gilvum_PYR-GCK GSALQGFTKLLGAPGEDGVFRYTPSPVD---DAGTRPED-DSDEVADWFS
Mvan_2759|M.vanbaalenii_PYR-1 GSALQGFTKLLGAPGEDGVFRYTPSPVD---ETSARPDD-DSDEVADWFS
Rv1488|M.tuberculosis_H37Rv NAALQGFTRLLGKPGEDGVFRFEPSPVE---DQPKHAADGDDAEVAGWFS
Mb1524|M.bovis_AF2122/97 NAALQGFTRLLGKPGEDGVFRFEPSPVE---DQPKHAADGDDAEVAGWFS
MMAR_2294|M.marinum_M NAALQGFTRMLGKPGEDGVFRFEASPVE---DLPKHATDGDDDEVSDWFS
MAV_3290|M.avium_104 SAALQGFTKLLGTPGQDGVFRFQPSPVE---DVPKHSAE-DDADVADWFS
MLBr_01802|M.leprae_Br4923 STALQGFTKLLGAPGEDGVFRFQPSPVDSSDQVVKHAADEDDAEVADWFS
MSMEG_3155|M.smegmatis_MC2_155 GTALQGFTKMLGAPGEDGVFRYTPSPVD---DTPPKTDD--DEEVADWFN
TH_1418|M.thermoresistible__bu GAALQGFTKLLGAPGEDGVFRFEPSPVE--EDLPRPEDD--SEEVADWFT
MAB_2718c|M.abscessus_ATCC_199 GSALQGFTKLLGAPGDDGVFRYEPSPVD---TSTERPED-DSDEVAGWFD
.:******::** **:*****: .***: : . :*:.**
Mflv_3653|M.gilvum_PYR-GCK TETDPVIAQAVAKAEAEAR--APVPGALDGPKQYPPLSQQVQPTATDYAQ
Mvan_2759|M.vanbaalenii_PYR-1 TETDPAIAQAVAKAEAEAR--APVAGTLDAPRQYPPLSQQAQPPATDYAQ
Rv1488|M.tuberculosis_H37Rv TDTDPSIARAVATAEAIAR--KPVEGSLGTPPRLTQ--------------
Mb1524|M.bovis_AF2122/97 TDTDPSIARAVATAEAIAR--KPVEGSLGTPPRLTQ--------------
MMAR_2294|M.marinum_M TETDPAIAQAVAKAEAIAR--KPVEGPLASPPELGPYNR-----------
MAV_3290|M.avium_104 TETDPAIAQAVAKAEAIAR--QPADGPTG---ELTQ--------------
MLBr_01802|M.leprae_Br4923 VETDPAIAQAVAKAEAMVY--QPVDE------------------------
MSMEG_3155|M.smegmatis_MC2_155 TETDPAIAEAVAKAEAEARTSTPTLGSSPATPPLGGTTPNPSINVPQTQE
TH_1418|M.thermoresistible__bu TQTDPAIAQAVAKAEAEAR--TPVDTGLTTPPLTERSRQQELP-------
MAB_2718c|M.abscessus_ATCC_199 TKATPEAAAAVAEAQSAAKEIGQIDPVSRHQLPEV---------------
..: * * *** *:: .
Mflv_3653|M.gilvum_PYR-GCK QP---RHGSPDQ-
Mvan_2759|M.vanbaalenii_PYR-1 QPPVPRHGAPDQ-
Rv1488|M.tuberculosis_H37Rv -------------
Mb1524|M.bovis_AF2122/97 -------------
MMAR_2294|M.marinum_M -------------
MAV_3290|M.avium_104 -------------
MLBr_01802|M.leprae_Br4923 -------------
MSMEG_3155|M.smegmatis_MC2_155 WPGGGAHHAQPEQ
TH_1418|M.thermoresistible__bu -------------
MAB_2718c|M.abscessus_ATCC_199 -------------