For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQFDAVLLLSFGGPEGPEQVRPFLENVTRGRGVPAERLDAVAEHYLHFGGVSPINGINRTLIAELEAQQE LPVYFGNRNWEPYVEDAVTAMRDNGVRRAAVFATSAWSGYSSCTQYVEDIARARRAAGRDAPELVKLRPY FDHPLFVEMFADAITAAAATVRGDARLVFTAHSIPTAADRRCGPNLYSRQVAYATRLVAAAAGYCDFDLA WQSRSGPPQVPWLEPDVTDQLTGLAGAGINAVIVCPIGFVADHIEVVWDLDHELRLQAEAAGIAYARAST PNADPRFARLARGLIDELRYGRIPARVSGPDPVPGCLSSINGQPCRPPHCVASVSPARPSAGSP
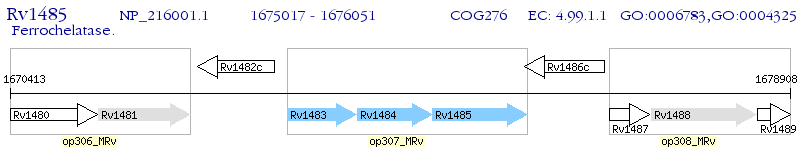
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1485 | hemH | - | 100% (344) | ferrochelatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1521 | hemH | 0.0 | 100.00% (344) | ferrochelatase |
| M. gilvum PYR-GCK | Mflv_3656 | hemH | 1e-138 | 73.65% (334) | ferrochelatase |
| M. leprae Br4923 | MLBr_01805 | hemH | 1e-152 | 76.79% (336) | ferrochelatase |
| M. abscessus ATCC 19977 | MAB_2721c | hemH | 1e-127 | 66.67% (333) | ferrochelatase |
| M. marinum M | MMAR_2291 | hemH | 1e-164 | 82.84% (338) | ferrochelatase HemZ |
| M. avium 104 | MAV_3293 | hemH | 1e-159 | 81.49% (335) | ferrochelatase |
| M. smegmatis MC2 155 | MSMEG_3152 | hemH | 1e-139 | 72.32% (336) | ferrochelatase |
| M. thermoresistible (build 8) | TH_1378 | hemZ | 1e-140 | 72.93% (351) | FERROCHELATASE HEMZ (PROTOHEME FERRO-LYASE) (HEME |
| M. ulcerans Agy99 | MUL_1493 | hemH | 1e-163 | 83.73% (332) | ferrochelatase |
| M. vanbaalenii PYR-1 | Mvan_2754 | hemH | 1e-135 | 72.37% (333) | ferrochelatase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3152|M.smegmatis_MC2_155 MSFDALLLLSFGGPEAPEQVMPFLENVTRGRGIPRERLESVAEHYLHFGG
Mvan_2754|M.vanbaalenii_PYR-1 MSVEALLLLSFGGPEGPEQVMPFLENVTRGRGIPRERLESVAEHYLHFGG
Mflv_3656|M.gilvum_PYR-GCK MTFDALLLLSFGGPEGPEQVMPFLENVTRGRGIPRERLESVAEHYLHFGG
Rv1485|M.tuberculosis_H37Rv MQFDAVLLLSFGGPEGPEQVRPFLENVTRGRGVPAERLDAVAEHYLHFGG
Mb1521|M.bovis_AF2122/97 MQFDAVLLLSFGGPEGPEQVRPFLENVTRGRGVPAERLDAVAEHYLHFGG
MMAR_2291|M.marinum_M MEFDAVLLLSFGGPEGPDQVRPFLENVTRGRGVPPERLDAVAEHYMHFGG
MUL_1493|M.ulcerans_Agy99 MEFDAVLLLSFGGPEGPEQVRPFLENVTRGRGVPPERLDAVAEHYMHFGG
MLBr_01805|M.leprae_Br4923 MLFDAALLLSFGGPDGPEQVRPFLENVTRGCNVPPERLDEVTKHYLHFGG
MAV_3293|M.avium_104 MDFDAVLLLSFGGPEGPEQVRPFLENVTRGRGVPPERLDHVAEHYLHFGG
TH_1378|M.thermoresistible__bu VEFDAVLLLSFGGPEGPEQVRPFLENVTRGRNVPPERLDDVAEHYLHFGG
MAB_2721c|M.abscessus_ATCC_199 MLFDAVLLLSFGGPEGPQEVRPFLENVTRGRGIPPERLDEVAEHYFHFGG
: .:* ********:.*::* ********* .:* ***: *::**:****
MSMEG_3152|M.smegmatis_MC2_155 VSPINGINRDLIVAIEAELARRGRNLPVYFGNRNWEPYVEDTVKAMSDNG
Mvan_2754|M.vanbaalenii_PYR-1 VSPINGINRDLIVAIEAELARRGMQMPVYFGNRNWEPYVEDTVAAMRDNG
Mflv_3656|M.gilvum_PYR-GCK VSPINGINRDLITAIEAELSRRGMQLPVYFGNRNWEPFVEDTVAAMRDNG
Rv1485|M.tuberculosis_H37Rv VSPINGINRTLIAELEAQQE-----LPVYFGNRNWEPYVEDAVTAMRDNG
Mb1521|M.bovis_AF2122/97 VSPINGINRTLIAELEAQQE-----LPVYFGNRNWEPYVEDAVTAMRDNG
MMAR_2291|M.marinum_M VSPINGINRALVTELRAEVG-----LPVYFGNRNWEPYVEDTVMAMRDNG
MUL_1493|M.ulcerans_Agy99 VSPINGINRALVTELRAEVG-----LPVYFGNRNWEPYVEDTVMAMRDNG
MLBr_01805|M.leprae_Br4923 VSPINGINLALVNELQVELD-----LPVYFGNRNWEPYIEDSVVTMRDDG
MAV_3293|M.avium_104 VSPINGINRALIEQLRAAQA-----LPVYFGNRNWEPYVEDTVKVMRDNG
TH_1378|M.thermoresistible__bu VSPLNAINRALITELEAELADRGLDLPVYFGNRNWEPYVEDTVTAMRDNG
MAB_2721c|M.abscessus_ATCC_199 VSPINGINRALVDQISRQLESAGHPLPVFFGNRNWHPMVEDTVAEMRDKG
***:*.** *: : :**:******.* :**:* * *.*
MSMEG_3152|M.smegmatis_MC2_155 IRRAAVFATSAWGGYSGCAQYQEDIARGRAAAGPEAPELVKLRQYFDHPL
Mvan_2754|M.vanbaalenii_PYR-1 IRRAAVFSTSAWGGYSGCAQYQEDIARGRAAAGPEAPELVKLRQYFDHPL
Mflv_3656|M.gilvum_PYR-GCK IRRAAVFSTSAWGGYSGCTQYQEDIARGRNAAGPDAPELVKLRQYFDHPL
Rv1485|M.tuberculosis_H37Rv VRRAAVFATSAWSGYSSCTQYVEDIARARRAAGRDAPELVKLRPYFDHPL
Mb1521|M.bovis_AF2122/97 VRRAAVFATSAWSGYSSCTQYVEDIARARRAAGRDAPELVKLRPYFDHPL
MMAR_2291|M.marinum_M VKRAAVFATSAWSGYSGCTQYVEDIARARRAAGPDAPELVKLRSYFDHPL
MUL_1493|M.ulcerans_Agy99 VKRAAVFATSAWSGYSGCTQYVEDIARARRAAGPDAPELVKLRSYFDHPL
MLBr_01805|M.leprae_Br4923 IRCAAVFITSAWSGYSSCTRYVEAIARARRRAGTGAPNLVKLRPYFDHPL
MAV_3293|M.avium_104 IRRAAVFTTSAWSGYSSCTQYVEDIARARAAAGPGAPELVKLRPYFDHPL
TH_1378|M.thermoresistible__bu IRRAAVFTTSAWGGYSSCTQYVEDIARGRRAAGPRAPELVKLRQYFDHPL
MAB_2721c|M.abscessus_ATCC_199 IRRAAVFTTSAWGGYSGCKQYVEDIARARAAVGDGAPELVKLRQYFDHPS
:: **** ****.***.* :* * ***.* .* **:***** *****
MSMEG_3152|M.smegmatis_MC2_155 FVEMFADAVADAAATLPEELRDEARLVFTAHSIPLRAASRCGADLYERQV
Mvan_2754|M.vanbaalenii_PYR-1 LIEMFADAIRDAAATLPEDLRAQARLVFTAHSIPVRAANRCGPDLYERQV
Mflv_3656|M.gilvum_PYR-GCK LVEMFADAIADARDTLPEPLRADARLVFTAHSIPLRAASRCGPDLYERQV
Rv1485|M.tuberculosis_H37Rv FVEMFADAITAAAATVR----GDARLVFTAHSIPTAADRRCGPNLYSRQV
Mb1521|M.bovis_AF2122/97 FVEMFADAITAAAATVR----GDARLVFTAHSIPTAADRRCGPNLYSRQV
MMAR_2291|M.marinum_M FVEMFADAIAAAATTVP----KDARLVFTAHSIPVAADRRCGPGLYSRQV
MUL_1493|M.ulcerans_Agy99 FVEMFADAIAAAAATVP----KGARLVFTAHSIPVAADRRCGPGLYSRQV
MLBr_01805|M.leprae_Br4923 FVEMFVDAITAAAASLPAALRSEARLVFTAHSVPVATDRRCGPALYSRQV
MAV_3293|M.avium_104 FVEMFAGAIADAAAKVP----AGARLVFTAHSVPVAADERVGPRLYSRQV
TH_1378|M.thermoresistible__bu LVEMFAEAIDTAAESLPAELRSQARLVFTAHSIPVAADEQHGPRLYSRQV
MAB_2721c|M.abscessus_ATCC_199 LVQLYAEAVSAAMSSLR----GEARLVFTAHSIPLAADRRHGPELYSRQV
:::::. *: * .: *********:* : : *. **.***
MSMEG_3152|M.smegmatis_MC2_155 GYAARLVAAAAGYREYDQVWQSRSGPPQVPWLEPDVGDHLEALARNGTRA
Mvan_2754|M.vanbaalenii_PYR-1 AHTSALVAAAAGYPEYDQVWQSRSGPPQVPWLEPDVGDHLEVLAARGVNA
Mflv_3656|M.gilvum_PYR-GCK GYAAGLVAGAAGYRDYDQVWQSRSGPPQVPWLEPDVGDHLAALAEAGTKA
Rv1485|M.tuberculosis_H37Rv AYATRLVAAAAGYCDFDLAWQSRSGPPQVPWLEPDVTDQLTGLAGAGINA
Mb1521|M.bovis_AF2122/97 AYATRLVAAAAGYCDFDLAWQSRSGPPQVPWLEPDVTDQLTGLAGAGINA
MMAR_2291|M.marinum_M AYAASLVAAAAGYDDYDLAWQSRSGPPQVPWLEPDVADHLETLRAGGVKA
MUL_1493|M.ulcerans_Agy99 AYAASLVAAAAGYDDYDLAWQSRSGPPQVPWLEPDVADHLEMLRAGGVKA
MLBr_01805|M.leprae_Br4923 GYAARLVAAGAGYADYDLTWQSRSGPPYVPWLAPDVGDQLMTLASAGTKA
MAV_3293|M.avium_104 AYAARLVAAAAGYAEHDLVWQSRSGPPQVRWLEPDVADHLRALAESGTPA
TH_1378|M.thermoresistible__bu RYAARLVAAAAGREHYDLVWQSRSGPPQIPWLEPDVGDHLGALAESGVRA
MAB_2721c|M.abscessus_ATCC_199 HYLAQLVAAATGHQDYDVVWQSRSGPPQVPWLEPDIVDHLAALKDRGIRS
: : ***..:* ..* .******** : ** **: *:* * * :
MSMEG_3152|M.smegmatis_MC2_155 VIVCPLGFVADHIEVVWDLDNELAEQAAEAGIAFARAATPNSQPRFAQLV
Mvan_2754|M.vanbaalenii_PYR-1 VIVCPVGFVADHIEVVWDLDNELAEQAAEAGIALARASTPNAQPRFAKLV
Mflv_3656|M.gilvum_PYR-GCK VIVCPIGFVSDHIEVVWDLDSELAEQAAEAGVALARASTPNAQPRFAQLA
Rv1485|M.tuberculosis_H37Rv VIVCPIGFVADHIEVVWDLDHELRLQAEAAGIAYARASTPNADPRFARLA
Mb1521|M.bovis_AF2122/97 VIVCPIGFVADHIEVVWDLDHELRLQAEAAGIAYARASTPNADPRFARLA
MMAR_2291|M.marinum_M VIVCPIGFVADHIEVVWDLDSELRAQAQGAGIALARAATPNADRRFARLA
MUL_1493|M.ulcerans_Agy99 VIVCPIGFVADHIEVVWDLDAELRAQAQDAGIALARAATPNADRRFARLA
MLBr_01805|M.leprae_Br4923 VIVCPIGFVADHIEVVWDLDHELRSQADAAGVAFARAATPNADRRFARLA
MAV_3293|M.avium_104 VIVCPIGFVADHIEVVWDLDEELRAQAESAGMLMARASTPNAQPRFARLA
TH_1378|M.thermoresistible__bu VIVCPIGFVADHIEVVYDLDHELGEQAAQAGIALARAGTPNSDRRFARLA
MAB_2721c|M.abscessus_ATCC_199 VAVAPIGFVSDHLEVVWDLDNEAREYAQEHDIELVRAATPGADERFARLA
* *.*:***:**:***:*** * * .: .**.**.:: ***:*.
MSMEG_3152|M.smegmatis_MC2_155 VDLIDEMLHGLPPRRVEGPDPVP------AYGSSVNGAPCT--PACSA--
Mvan_2754|M.vanbaalenii_PYR-1 VDLIDELRLGLPPQRVGG-GLVP------GYGSSVNGALCT--PDCSG--
Mflv_3656|M.gilvum_PYR-GCK VDLIDEVIAGRPPQRVPGENPVP------GYGSSVNGALCT--ELCGA--
Rv1485|M.tuberculosis_H37Rv RGLIDELRYGRIPARVSGPDPVP------GCLSSINGQPCR-PPHCVASV
Mb1521|M.bovis_AF2122/97 RGLIDELRYGRIPARVSGPDPVP------GCLSSINGQPCR-PPHCVASV
MMAR_2291|M.marinum_M RGLIEELRSGGEPARVGGPDPVF------GCLAGINGEPCR-PPHCAARE
MUL_1493|M.ulcerans_Agy99 RGLIEELRSGGEPARVGGPDSVS------GCLAGVNGEPCR-PPHCAARE
MLBr_01805|M.leprae_Br4923 ASLIDELTHDRVPVRVNGSDPVP------GCLASINGVPCD-LPHCVA--
MAV_3293|M.avium_104 ADLIDELRCGRTPARVTGPDPVP------GCLASVNGAPCR-PPHCAAQA
TH_1378|M.thermoresistible__bu VDLIDELRLGLEPARVSGPAALPAEFRAPGCGHVVDGTPCAAAPNCVSVI
MAB_2721c|M.abscessus_ATCC_199 VDLITEVVEGRPALRASGVNPVP------GYGFSVNGALCS--PLCCGVQ
.** *: . . *. * : . ::* * * .
MSMEG_3152|M.smegmatis_MC2_155 ----------
Mvan_2754|M.vanbaalenii_PYR-1 ----------
Mflv_3656|M.gilvum_PYR-GCK ----------
Rv1485|M.tuberculosis_H37Rv SPARPSAGSP
Mb1521|M.bovis_AF2122/97 SPARPSAGSP
MMAR_2291|M.marinum_M TAAQGA----
MUL_1493|M.ulcerans_Agy99 TAPQGA----
MLBr_01805|M.leprae_Br4923 ----------
MAV_3293|M.avium_104 TG--------
TH_1378|M.thermoresistible__bu ATRTR-----
MAB_2721c|M.abscessus_ATCC_199 ED--------