For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MACLGRPGCRGWAGASLVLVVVLALAACTESVAGRAMRATDRSSGLPTSAKPARARDLLLQDGDRAPFGQ VTQSRVGDSYFTSAVPPECSAALLFKGSPLRPDGSSDHAEAAYNVTGPLPYAESVDVYTNVLNVHDVVWN GFRDVSHCRGDAVGVSRAGRSTPMRLRYFATLSDGVLVWTMSNPRWTCDYGLAVVPHAVLVLSACGFKPG FPMAEWASKRRAQLDSQV
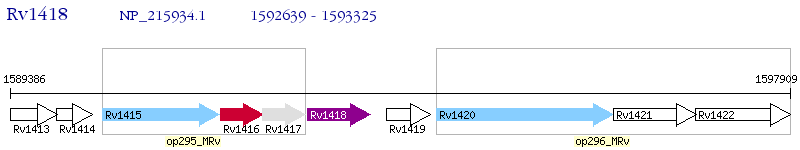
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1418 | lprH | - | 100% (228) | PROBABLE LIPOPROTEIN LPRH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1453 | lprH | 1e-133 | 100.00% (228) | lipoprotein LprH |
| M. gilvum PYR-GCK | Mflv_3443 | - | 7e-06 | 25.23% (222) | hypothetical protein Mflv_3443 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2225 | lprH | 2e-67 | 61.21% (214) | lipoprotein LprH |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_4018 | - | 4e-05 | 20.00% (190) | PUTATIVE putative hypothetical protein Mflv_3443 |
| M. ulcerans Agy99 | MUL_1812 | lprH | 2e-67 | 61.32% (212) | lipoprotein LprH |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1418|M.tuberculosis_H37Rv MACLGRPGCRGWAGASLVLVVVLALAACTESVAGRAMRATDRSSGLPTSA
Mb1453|M.bovis_AF2122/97 MACLGRPGCRGWAGASLVLVVVLALAACTESVAGRAMRATDRSSGLPTSA
MMAR_2225|M.marinum_M MTRLGRS-----CAAVVLLVVVVAMAACTNSVAGRAIRAS----GQPR--
MUL_1812|M.ulcerans_Agy99 ---------------MVLLVVVVAMAACTNSVAGRAIRAS----GQPR--
Mflv_3443|M.gilvum_PYR-GCK --MTIAARRRTAAAASAVLCTAALLAGCTRLVDDPQARP-----------
TH_4018|M.thermoresistible__bu --MRLTPTRLTRAGAVAALL-AVVACGCTRTLDTATPDPG----------
* . ..**. : .
Rv1418|M.tuberculosis_H37Rv KPARARDLLLQDGDRAPFGQVTQSRVGDSYFTSAVPPECSAALLFKGSPL
Mb1453|M.bovis_AF2122/97 KPARARDLLLQDGDRAPFGQVTQSRVGDSYFTSAVPPECSAALLFKGSPL
MMAR_2225|M.marinum_M AHILARDLLLQDGDNTPLGPASSTRVGETYFTSARPPECTAAVLFKGSPL
MUL_1812|M.ulcerans_Agy99 AHISARDLLLQDGDNTPLGPASSTRVGETYFTSARPPECTAAVLFKGSPL
Mflv_3443|M.gilvum_PYR-GCK ---QALAAPITELQVVDLLSPEVVGEDGNLFVVAEPQRCAGLAREVDPPL
TH_4018|M.thermoresistible__bu ---GPPVAPITAGQVGDLLSPEVAGGEGSLFTTVEPEDCAGVAREVDPPF
. : : : . *. . * *:. ..*:
Rv1418|M.tuberculosis_H37Rv RPDGSSDHAEAAYN---VTGPLPYAESVDVYTNVLNVHDVVWNGFRDVSH
Mb1453|M.bovis_AF2122/97 RPDGSSDHAEAAYN---VTGPLPYAESVDVYTNVLNVHDVVWNGFRDVSH
MMAR_2225|M.marinum_M RPPGSADFAESAYR---VQATAMYAESVDVYNSPLNPHDVVQNGFRAVSQ
MUL_1812|M.ulcerans_Agy99 RPPGSADFAESAYR---VQATAMYAESVDVYNSPLNPHDVVQNGFRVVSQ
Mflv_3443|M.gilvum_PYR-GCK IEAGRPVATDGGHWTSEDGR-FYIEEMVAVYLSDFDAEGALRSARATVDE
TH_4018|M.thermoresistible__bu IEDFDPAATTGGHWRDDVGGGVVIEEMVGVYRADFDPQEALARARQTIDS
. : ..: * * ** :: . .: . :.
Rv1418|M.tuberculosis_H37Rv CRGDAVGVSRAG--RSTPMRLRYFATLSDGVLVWTMSNPRWTCDYGLAVV
Mb1453|M.bovis_AF2122/97 CRGDAVGVSRAG--RSTPMRLRYFATLSDGVLVWTMSNPRWTCDYGLAVV
MMAR_2225|M.marinum_M CHGDAFAGYPAG--EFGPITLKSFGTPAEGVLVWSMGRSDWNCDYGLAVV
MUL_1812|M.ulcerans_Agy99 CHGDAFAGYPAG--EFGPITLKSFGTPAEGVLVWSMGRSDWNCDYGLAVV
Mflv_3443|M.gilvum_PYR-GCK CLGSRLSVTTMRDRVYDFEVAPGAE-GPDGSVLWSLRAVDWNCDNLFVAA
TH_4018|M.thermoresistible__bu CRDVPITVTAMSGDTYDFRLLPPVDSGSPDILVWAFRADNWACDNTLVAA
* . . . . ::*:: * ** :...
Rv1418|M.tuberculosis_H37Rv PHAVLVLSACGFKPGFPMAEWASKRRAQLDSQV----
Mb1453|M.bovis_AF2122/97 PHAVLVLSACGFKPGFPMAEWASKRRAQLDSQV----
MMAR_2225|M.marinum_M PRAALVISACDQNSGFPMAEWAVTRRAQLDDRV----
MUL_1812|M.ulcerans_Agy99 PRAALVISACDQNSGFPMAEWAVTRRAQLDDRV----
Mflv_3443|M.gilvum_PYR-GCK YNAAVEITTCGAAPGFDVAALAADALERIRRLADTTA
TH_4018|M.thermoresistible__bu HNAAVEITTCSRTGGYDVLSLAQQALKRIEALANTTV
.*.: :::*. *: : * :: .