For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRVRLVIAQCTVDYIGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPLNWMSPPCWLTEESGGQAPVW VVENKAGEQLRITIEGIEHDSSHELGVDPGLVKDGVEAHLQALLAEHIQLLGEGYTLVRREYMTAIGPVD LLCSDERGGSVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVKGVFAAQQIKPQARILATDRGIRCLTL DYDTMRGMDSGEYRLF
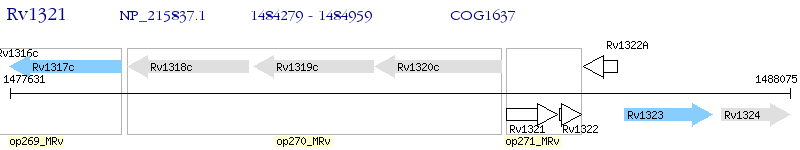
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1321 | - | - | 100% (226) | hypothetical protein Rv1321 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1355 | - | 1e-128 | 99.56% (226) | hypothetical protein Mb1355 |
| M. gilvum PYR-GCK | Mflv_2337 | - | 1e-114 | 89.69% (223) | hypothetical protein Mflv_2337 |
| M. leprae Br4923 | MLBr_01155 | - | 1e-112 | 88.18% (220) | hypothetical protein MLBr_01155 |
| M. abscessus ATCC 19977 | MAB_1460 | - | 1e-109 | 83.19% (232) | hypothetical protein MAB_1460 |
| M. marinum M | MMAR_4077 | - | 1e-118 | 91.93% (223) | hypothetical protein MMAR_4077 |
| M. avium 104 | MAV_1541 | - | 1e-118 | 92.73% (220) | hypothetical protein MAV_1541 |
| M. smegmatis MC2 155 | MSMEG_4923 | - | 1e-110 | 87.05% (224) | hypothetical protein MSMEG_4923 |
| M. thermoresistible (build 8) | TH_1536 | - | 1e-114 | 87.44% (223) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3940 | - | 1e-119 | 91.59% (226) | hypothetical protein MUL_3940 |
| M. vanbaalenii PYR-1 | Mvan_4308 | - | 1e-114 | 89.24% (223) | hypothetical protein Mvan_4308 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2337|M.gilvum_PYR-GCK ---MRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
Mvan_4308|M.vanbaalenii_PYR-1 ---MRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MSMEG_4923|M.smegmatis_MC2_155 ---MRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
Rv1321|M.tuberculosis_H37Rv MSRVRLVIAQCTVDYIGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
Mb1355|M.bovis_AF2122/97 MSRVRLVIAQCTVDYIGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MMAR_4077|M.marinum_M ---MRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MUL_3940|M.ulcerans_Agy99 MIRVRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MLBr_01155|M.leprae_Br4923 ------MIAQCTVDYLGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MAV_1541|M.avium_104 ------MIAQCTVDYVGRLTAHLPSARRLLLFKVDGSVSVHADDRAYKPL
TH_1536|M.thermoresistible__bu ---VRLVIAQCTVDYVGRLTAHLPSARRLLLFKADGSVSVHADDRAYKPL
MAB_1460|M.abscessus_ATCC_1997 ---MRLVVAQCTVNYVGRLTAHLPSAKRLLLIKADGSVSVHADDRAYKPL
::*****:*:**********:****:*.****************
Mflv_2337|M.gilvum_PYR-GCK NWMSPPCRLVEQ---------ADGETPVWVVENKAGEQLRITVEAIEHDS
Mvan_4308|M.vanbaalenii_PYR-1 NWMSPPCWIVEP---------TDGDATVWVVENKAGEQLRITVEAIEHDS
MSMEG_4923|M.smegmatis_MC2_155 NWMSPPCWVTEQD--------TETGVALWVVENKTGEQLRITVEDIEHDS
Rv1321|M.tuberculosis_H37Rv NWMSPPCWLTEE---------SGGQAPVWVVENKAGEQLRITIEGIEHDS
Mb1355|M.bovis_AF2122/97 NWMSPPCWLTEE---------SGGQAPVWVVENKAGEQLRITIEGIEHDS
MMAR_4077|M.marinum_M NWMSPPCWLTEE---------PGGDSPVWVVTNKGGEQLRISVEEIEHDS
MUL_3940|M.ulcerans_Agy99 NWMSPPCWLTEE---------PGGDSPVWVVTNKGGEQLRISVEEIEHDS
MLBr_01155|M.leprae_Br4923 NWMSPPCWLTED---------VAGASPAWVVENKAGEQLRITVEEVEHDS
MAV_1541|M.avium_104 NWMSPPCWLREE---------AGDAAPVWVVENKAGEQLRITVEDIEHDS
TH_1536|M.thermoresistible__bu NWMSPPCWLTEH---------NDGEVPVWVVENKAGEQLRITVENIDHDS
MAB_1460|M.abscessus_ATCC_1997 NWMSPPCWLTETEGEGGDDTSESGAPTVWVVENKAGEQLRITIESIEHDS
******* : * . *** ** ******::* ::***
Mflv_2337|M.gilvum_PYR-GCK SHELGVDPGLVKDGVEAHLQALLAEHVELLGAGYTLVRREYMTPIGPVDL
Mvan_4308|M.vanbaalenii_PYR-1 SHELGVDPGLVKDGVEAHLQALLAEHVELLGAGYTLVRREYMTPIGPVDL
MSMEG_4923|M.smegmatis_MC2_155 HHELGVDPGLVKDGVEAHLQALLAEHVELLGAGYTLVRREYPTPIGPVDL
Rv1321|M.tuberculosis_H37Rv SHELGVDPGLVKDGVEAHLQALLAEHIQLLGEGYTLVRREYMTAIGPVDL
Mb1355|M.bovis_AF2122/97 SHELGVDPGLVKDGVEAHLQALLAEHIQLLGEGYTLVRREYMTAIGPVDL
MMAR_4077|M.marinum_M SHELGVDPGLVKDGVEAHLQVLLAEHVQLLGEGYTLVRREYMTAIGPVDL
MUL_3940|M.ulcerans_Agy99 SHELGVDPGLVKDGVEAHLQVLLAEHVQLLGEGYTLVRREYMTAIGPVDL
MLBr_01155|M.leprae_Br4923 CHDLGVDPGLVKDGVEAHLQTLLAEQVQLLGEGYTLVRCEYMTAIGPVDL
MAV_1541|M.avium_104 SHDLGVDPGLVKDGVEAHLQALLAEHVQLLGEGYTLVRREYMTAIGPVDL
TH_1536|M.thermoresistible__bu THDLGVDPGLVKDGVEAHLQELLAEHVELLGPGYTLVRREYMTAIGPVDL
MAB_1460|M.abscessus_ATCC_1997 AHELGVDPGLVKDGVEAHLQELLAEHVALLGDGYTLVRREYMTPIGPVDL
*:***************** ****:: *** ****** ** *.******
Mflv_2337|M.gilvum_PYR-GCK LCRDEQGRSVAVEIKRRGEIDGVEQLTRYLELLNRDSLLAPVAGVFAAQQ
Mvan_4308|M.vanbaalenii_PYR-1 LCRDEQGRSVAVEIKRRGEIDGVEQLTRYLELLNRDSLLAPVAGVFAAQQ
MSMEG_4923|M.smegmatis_MC2_155 LCRDELGRSVAVEIKRRGEIDGVEQLTRYLELLNRDSLLAPVAGVFAAQQ
Rv1321|M.tuberculosis_H37Rv LCSDERGGSVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVKGVFAAQQ
Mb1355|M.bovis_AF2122/97 LCRDERGGSVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVKGVFAAQQ
MMAR_4077|M.marinum_M LCRDESGGAVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVKGVFAAQQ
MUL_3940|M.ulcerans_Agy99 LCRDESGGAVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVKGVFAAQQ
MLBr_01155|M.leprae_Br4923 LCRDEQGGSVAVEIKRRGEIDGVEQLTRYLALLNRNSVLAPVRGVFAAQH
MAV_1541|M.avium_104 LCRDERGGSVAVEIKRRGEIDGVEQLTRYLELLNRDSVLAPVRGVFAAQQ
TH_1536|M.thermoresistible__bu LCRDGNGGTVAVEIKRRGEIDGVEQLTRYLDLLNRDSLLAPVSGIFAAQQ
MAB_1460|M.abscessus_ATCC_1997 LCRDADGATVAVEIKRRGEIDGVEQLTRYLELLNRDTTLAPVAGVFAAQQ
** * * :********************* ****:: **** *:****:
Mflv_2337|M.gilvum_PYR-GCK IKPQARTLATDRGIRCLTLDYDKMRGMDSDEFRLF
Mvan_4308|M.vanbaalenii_PYR-1 IKPQARTLATDRGIRCLTLDYDKMRGMDNDEFRLF
MSMEG_4923|M.smegmatis_MC2_155 IKPQARTLATDRGIRCVTLDYDQMRGMDSDEYRLF
Rv1321|M.tuberculosis_H37Rv IKPQARILATDRGIRCLTLDYDTMRGMDSGEYRLF
Mb1355|M.bovis_AF2122/97 IKPQARILATDRGIRCLTLDYDTMRGMDSGEYRLF
MMAR_4077|M.marinum_M IKPQARTLATDRGIRCVTLDYDKMRGMDSDEYRLF
MUL_3940|M.ulcerans_Agy99 IKPQARTLATDRGIRCVTLDYDKMRGMDSDEYRLF
MLBr_01155|M.leprae_Br4923 IKPQARTLATDRGIRCVTLDYDKMRGMDSDEYRLF
MAV_1541|M.avium_104 IKPQARTLATDRGIRCVTLDYDKMRGMDSDEYRLF
TH_1536|M.thermoresistible__bu IKPQARTLAEDRGIRCVTLDYDKMRGMDSDEYRLF
MAB_1460|M.abscessus_ATCC_1997 IKPQARTLAEDRGIRCLTLDYDAMRGMDSDEFRLF
****** ** ******:***** *****..*:***