For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIIPDINLLLYAVITGFPQHRRAHAWWQDTVNGHTRIGLTYPALFGFLRIATSARVLAAPLPTADAIAYV REWLSQPNVDLLTAGPRHLDIALGLLDKLGTASHLTTDVQLAAYGIEYDAEIHSSDTDFARFADLKWTDP LRE
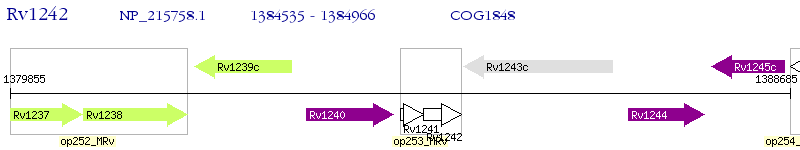
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1242 | - | - | 100% (143) | hypothetical protein Rv1242 |
| M. tuberculosis H37Rv | Rv2103c | - | 1e-23 | 37.68% (138) | hypothetical protein Rv2103c |
| M. tuberculosis H37Rv | Rv0277c | - | 2e-15 | 32.35% (136) | hypothetical protein Rv0277c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1274 | - | 3e-81 | 100.00% (143) | hypothetical protein Mb1274 |
| M. gilvum PYR-GCK | Mflv_0928 | - | 1e-21 | 34.06% (138) | PilT domain-containing protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1242|M.tuberculosis_H37Rv MIIPDINLLLYAVITGFPQHRRAHAWWQDTVNGHTRIGLTYPALFGFLRI
Mb1274|M.bovis_AF2122/97 MIIPDINLLLYAVITGFPQHRRAHAWWQDTVNGHTRIGLTYPALFGFLRI
Mflv_0928|M.gilvum_PYR-GCK MRIVDANVLLYAVNSSSEHHDASRRWLDGALSGNDVVGLAWVPMLAFVRL
* * * *:***** :. :* :: * :.::.*: :**:: .::.*:*:
Rv1242|M.tuberculosis_H37Rv ATSARVLAAPLPTADAIAYVREWLSQPNVDLLTAGPRHLDIALGLLDKLG
Mb1274|M.bovis_AF2122/97 ATSARVLAAPLPTADAIAYVREWLSQPNVDLLTAGPRHLDIALGLLDKLG
Mflv_0928|M.gilvum_PYR-GCK TTKHGLFPSPLTPDDAMARITDWCSAPGAVIVNPTARHADVLATLLSDVG
:*. ::.:**.. **:* : :* * *.. ::.. .** *: **..:*
Rv1242|M.tuberculosis_H37Rv TASHLTTDVQLAAYGIEYDAEIHSSDTDFARFADLKWTDPLRE--
Mb1274|M.bovis_AF2122/97 TASHLTTDVQLAAYGIEYDAEIHSSDTDFARFADLKWTDPLRE--
Mflv_0928|M.gilvum_PYR-GCK TGGNLVNDAHLAALALEHRARIVSYDNDFGRFSGVQWDVPESLLG
*..:*..*.:*** .:*: *.* * *.**.**:.::* *