For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VALVLVYLVVLVLVAIVLFAAASLLFGRGEQLPPLPRATTATTLPAFGVTRADVDAVKFTQVLRGYKTSE VDWVLERLGRELEALRSQLGAIHASSEDAEAESDASNPSRGETVVHYRSDPA
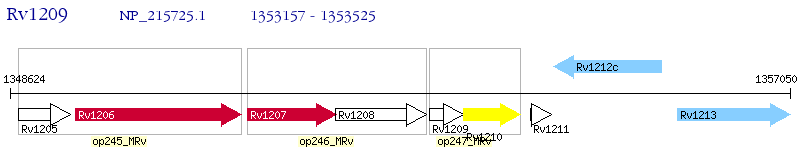
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1209 | - | - | 100% (122) | hypothetical protein Rv1209 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1241 | - | 1e-63 | 100.00% (122) | hypothetical protein Mb1241 |
| M. gilvum PYR-GCK | Mflv_2188 | - | 2e-36 | 63.33% (120) | hypothetical protein Mflv_2188 |
| M. leprae Br4923 | MLBr_01065 | - | 6e-43 | 79.46% (112) | hypothetical protein MLBr_01065 |
| M. abscessus ATCC 19977 | MAB_1348 | - | 3e-30 | 57.69% (104) | hypothetical protein MAB_1348 |
| M. marinum M | MMAR_4229 | - | 8e-43 | 73.95% (119) | hypothetical protein MMAR_4229 |
| M. avium 104 | MAV_1354 | - | 1e-42 | 75.68% (111) | hypothetical protein MAV_1354 |
| M. smegmatis MC2 155 | MSMEG_5083 | - | 4e-34 | 61.34% (119) | hypothetical protein MSMEG_5083 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0960 | - | 5e-43 | 73.95% (119) | hypothetical protein MUL_0960 |
| M. vanbaalenii PYR-1 | Mvan_4507 | - | 2e-36 | 64.46% (121) | hypothetical protein Mvan_4507 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2188|M.gilvum_PYR-GCK MTLVLLYLVVLVLVGIVLFAVGSVLFGRGEVLPPLPQGTTATVLPASGVT
Mvan_4507|M.vanbaalenii_PYR-1 MTLVLLYLVVLVLVGIVLFAVGSVLFGRGEVLPPLPQATTAAVLPASGVT
MSMEG_5083|M.smegmatis_MC2_155 MTLILLYLVVLVLVATVLFALGSVIFGRGETLPPLPRGTTATVLPASGVT
Rv1209|M.tuberculosis_H37Rv MALVLVYLVVLVLVAIVLFAAASLLFGRGEQLPPLPRATTATTLPAFGVT
Mb1241|M.bovis_AF2122/97 MALVLVYLVVLVLVAIVLFAAASLLFGRGEQLPPLPRATTATTLPAFGVT
MMAR_4229|M.marinum_M MALVLLYLVVLVLVAVVLFGVASLLFGRGEQLPPLPRATTSTVLPAYGVT
MUL_0960|M.ulcerans_Agy99 MALVLLYLVVLVLVAVVLFGVASLLFGRGEQLPPLPRATTSTVLPAYGVT
MAV_1354|M.avium_104 MALVLLYLVVLVLVAIVLFGAASLLFGRGEQLPPLPRGTTATVLPAYGVT
MLBr_01065|M.leprae_Br4923 MALVLLYLVVLVLVAIVLFGAASLLFGRGERLPPLPRGTTATVLPAHGVT
MAB_1348|M.abscessus_ATCC_1997 MTFALTYLLVLIAVAAVLFVLGSMLFGRGEQLPPLPKGTTATVLPADKVT
*:: * **:**: *. *** .*::***** *****:.**::.*** **
Mflv_2188|M.gilvum_PYR-GCK GADVDAVKFTQTLRGYKTGEVDWVLDRLGREIDALRGELAAVRAAHGVED
Mvan_4507|M.vanbaalenii_PYR-1 GADVEAVKFTQVLRGYKTSEVDWVLDRLGRELDMLRGELATVRAAHGLDG
MSMEG_5083|M.smegmatis_MC2_155 GADVESVRFTQSLRGYKTSEVDWVLERLGYELDSLRGELAALRVAYGVPE
Rv1209|M.tuberculosis_H37Rv RADVDAVKFTQVLRGYKTSEVDWVLERLGRELEALRSQLGAIHASSE-DA
Mb1241|M.bovis_AF2122/97 RADVDAVKFTQVLRGYKTSEVDWVLERLGRELEALRSQLGAIHASSE-DA
MMAR_4229|M.marinum_M RADVDAVKFTQVLRGYKTSEVDWVLDRLGRELEELREQLAAVHTSSTSES
MUL_0960|M.ulcerans_Agy99 RADVDAVKFTQVLRGYKTSEVDWVLDRLGRELEELREQLAAVHTSSTSES
MAV_1354|M.avium_104 GSDVDAVKFTQVLRGYKTSEVDWVLDRLARELEALRGQLAAAHAAAG-EQ
MLBr_01065|M.leprae_Br4923 GADVDAVKFTQVLRGYKPSEVDWVLDRLGRELEALRGQLAAIA-----DA
MAB_1348|M.abscessus_ATCC_1997 GADVDAVKFSLVFRGYKASEVDWVLDRLARQIDDLRAELDEVR-----DA
:**::*:*: :****..******:**. ::: ** :*
Mflv_2188|M.gilvum_PYR-GCK PTLDDAQS----SDAHPGEYARDDM--
Mvan_4507|M.vanbaalenii_PYR-1 PAEGDGPDG---AHALPAEQTRGDA--
MSMEG_5083|M.smegmatis_MC2_155 DTPEDIEDDDVEVSATAGELRGEEGTS
Rv1209|M.tuberculosis_H37Rv EAESDASNP---SRGETVVHYRSDPA-
Mb1241|M.bovis_AF2122/97 EAESDASNP---SRGETVVHYRSDPA-
MMAR_4229|M.marinum_M PAGPEDSDA---PGSQPAVSNGADST-
MUL_0960|M.ulcerans_Agy99 PAGPEDSDA---PGSQPAVSNGADST-
MAV_1354|M.avium_104 QTGGQSAEP---DDG----ADRQD---
MLBr_01065|M.leprae_Br4923 EADADVSNV---PSG----DDGQDVT-
MAB_1348|M.abscessus_ATCC_1997 RDNVDANAE------------------
: