For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVDAHTHLDACGARDADTVRSLVERAAAAGVTAVVTVADDLESARWVTRAAEWDRRVYAAVALHPTRADA LTDAARAELERLVAHPRVVAVGETGIDMYWPGRLDGCAEPHVQREAFAWHIDLAKRTGKPLMIHNRQADR DVLDVLRAEGAPDTVILHCFSSDAAMARTCVDAGWLLSLSGTVSFRTARELREAVPLMPVEQLLVETDAP YLTPHPHRGLANEPYCLPYTVRALAELVNRRPEEVALITTSNARRAYGLGWMRQ
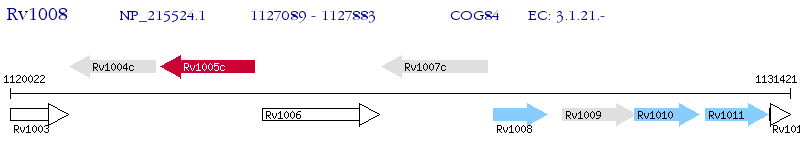
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1008 | tatD | - | 100% (264) | PROBABLE DEOXYRIBONUCLEASE TATD (YJJV PROTEIN) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1035 | tatD | 1e-152 | 99.62% (264) | deoxyribonuclease TatD (YjjV protein) |
| M. gilvum PYR-GCK | Mflv_1931 | - | 1e-117 | 77.22% (259) | TatD family hydrolase |
| M. leprae Br4923 | MLBr_00239 | - | 1e-126 | 82.69% (260) | hypothetical protein MLBr_00239 |
| M. abscessus ATCC 19977 | MAB_1129 | - | 1e-106 | 72.33% (253) | deoxyribonuclease TatD |
| M. marinum M | MMAR_4480 | tatD | 1e-127 | 83.85% (260) | deoxyribonuclease TatD |
| M. avium 104 | MAV_1146 | - | 1e-125 | 83.21% (262) | hydrolase, TatD family protein |
| M. smegmatis MC2 155 | MSMEG_5440 | - | 1e-111 | 72.97% (259) | deoxyribonuclease |
| M. thermoresistible (build 8) | TH_0978 | - | 2e-77 | 77.33% (172) | PROBABLE DEOXYRIBONUCLEASE TATD (YJJV PROTEIN) |
| M. ulcerans Agy99 | MUL_4653 | tatD | 1e-125 | 83.08% (260) | deoxyribonuclease TatD |
| M. vanbaalenii PYR-1 | Mvan_4802 | - | 1e-117 | 76.06% (259) | TatD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1931|M.gilvum_PYR-GCK -----MVVSADKQRRVPPPVPEPLDPLVDAHTHLDACGAD---DADGVRA
Mvan_4802|M.vanbaalenii_PYR-1 -------MSATKQRREPPPIPQPLAPLIDAHTHLDACGAR---NADDVRA
MSMEG_5440|M.smegmatis_MC2_155 -------MSAKRSAREKPPAPEPLTPLVDAHTHLDACGAD---NPEDVRA
TH_0978|M.thermoresistible__bu --------------------------------------------------
Rv1008|M.tuberculosis_H37Rv --------------------------MVDAHTHLDACGAR---DADTVRS
Mb1035|M.bovis_AF2122/97 --------------------------MVDAHTHLDACGAR---DADTVRS
MMAR_4480|M.marinum_M ----MRVSSNRAAEREAPPAPQPLAPLIDAHTHLDACGAA---DAAGVIA
MUL_4653|M.ulcerans_Agy99 --------------------------MIDAHTHLDACGAA---DAAGVIA
MLBr_00239|M.leprae_Br4923 --------------------------MVDAHTHLDACGAR---DADDVNA
MAV_1146|M.avium_104 ----MRVSSNRR-ERQAPPAPEPLAPLIDAHTHLDACAAGPETGAADVRA
MAB_1129|M.abscessus_ATCC_1997 MTPGNNRQAPPDRIRSRPPDPEPLAPLIDAHTHLDACGAV---TAADVRA
Mflv_1931|M.gilvum_PYR-GCK ILDRAGSAGVRAVVTIADDLESARWAVEAADWDSRVYAAVALHPTRAGAL
Mvan_4802|M.vanbaalenii_PYR-1 ILDRAASAGVQAVVTIADDLEAARWAAEAADWDPRVYAAVALHPTRANAL
MSMEG_5440|M.smegmatis_MC2_155 IVDRAAAVGVGRVVTIADDLDAAHWVTRAADADDRVWAAVALHPTRADTL
TH_0978|M.thermoresistible__bu --------------------------------------------------
Rv1008|M.tuberculosis_H37Rv LVERAAAAGVTAVVTVADDLESARWVTRAAEWDRRVYAAVALHPTRADAL
Mb1035|M.bovis_AF2122/97 LVERAAAAGVTAVVTVADDLESARWVTRAAEWDRRVYAAVALHPTRADAL
MMAR_4480|M.marinum_M IVDRAAAVGVEAVVTIADDLESARWVTRAAEWDPRVYAAVALHPTRTDAL
MUL_4653|M.ulcerans_Agy99 IVHRAAAVGVEAVVTIADDLESARWVTRAAEWDPRVYAAVALHPTRTDAL
MLBr_00239|M.leprae_Br4923 IVERAAAVGVVAVVTVADDLESARWVIAAAGWDRRVYAAVALHPTRANSL
MAV_1146|M.avium_104 IVDRAAAVGVQAVVTVADDLDSARWVTRAAEWDPRVYAATALHPTRADAL
MAB_1129|M.abscessus_ATCC_1997 MVDHAESVGVGRVVTVADDLDSARWAVSAAAADPRVYAAVALHPTRAGNL
Mflv_1931|M.gilvum_PYR-GCK TDEARTVLERLASHPRVVAVGETGMDLYWPGRLEGCAEPAEQREAFAWHI
Mvan_4802|M.vanbaalenii_PYR-1 TPEARSVIERLAAHPRVVAVGETGMDLYWPGRLEGCAEPADQREAFAWHI
MSMEG_5440|M.smegmatis_MC2_155 TDAAKADLEQLATHPRVVAIGETGMDLYWPGRLDGCATPAEQRDAFAWHI
TH_0978|M.thermoresistible__bu ----------------VVAIGETGMDLYWPGRLDGCADPAAQREAFAWHI
Rv1008|M.tuberculosis_H37Rv TDAARAELERLVAHPRVVAVGETGIDMYWPGRLDGCAEPHVQREAFAWHI
Mb1035|M.bovis_AF2122/97 TDAARAELERLVAHPRVVAVGETGIDMYWPGRLDGCAEPHVQREAFAWHI
MMAR_4480|M.marinum_M TEAARREIEELVAHRRVVAVGETGMDMYWPGRLEGCADPDVQREAFAWHI
MUL_4653|M.ulcerans_Agy99 IEAARREIEELVAHRRVVAVGETGMDMYWPGRLEGCADPDVQREAFAWHI
MLBr_00239|M.leprae_Br4923 TSAARAKIEQLAAHPRMVAVGETGMDLYWPGRLDSCAPPDVQREAFAWHI
MAV_1146|M.avium_104 DDAARAEIERLVAHPRVVAVGETGMDLYWPGRLDGCAEPAVQREAFAWHI
MAB_1129|M.abscessus_ATCC_1997 TDEARAELETLAAHPRVVAIGETGLDLYWPGRLEGCAEPSIQREAFAWHI
:**:****:*:******:.** * **:******
Mflv_1931|M.gilvum_PYR-GCK DLAKRTGKPLMIHNRDADAAVLDVLRAEGAPETVIFHCFSSDATMARACI
Mvan_4802|M.vanbaalenii_PYR-1 DLAKRTGKPLMIHNRDADAAVLDVLRAEGAPETVIFHCFSSGAAMARACI
MSMEG_5440|M.smegmatis_MC2_155 DLAKRSGKPLMIHNREADAEVLDVLRAEGAPETVIFHCFSSGPAMARTCI
TH_0978|M.thermoresistible__bu DLAKRTGKPLMIHNREADAEVLDVLRAEGAPETVIMHCFSSDAVMARTCV
Rv1008|M.tuberculosis_H37Rv DLAKRTGKPLMIHNRQADRDVLDVLRAEGAPDTVILHCFSSDAAMARTCV
Mb1035|M.bovis_AF2122/97 DLAKRTGKPLMIHNRQADRDVLDVLRAEGAPDTVILHCFSSDAAMARTCV
MMAR_4480|M.marinum_M DLAKRTGKPLMIHNRQADREVLDVLRAEGAPDTVIFHCFSSDAAMARECV
MUL_4653|M.ulcerans_Agy99 DLAKRTGKPLMIHNRQADREVLDVLRAEGAPDTVIFHCFSSDAAMARECV
MLBr_00239|M.leprae_Br4923 DLAKRCGKPLMIHNRQADAEVLDVLRAEGAPEQVIFHCFSSNAAMARRCV
MAV_1146|M.avium_104 DLAKRCGKPLMIHNREADAEVLDVLAAEGAPDVVIFHCFSSDAAMARRCV
MAB_1129|M.abscessus_ATCC_1997 ELAKRTGKPLMIHNRDADADVLDVLKAEGAPGSVIFHCFSSDADMAKKCI
:**** *********:** ***** ***** **:*****.. **: *:
Mflv_1931|M.gilvum_PYR-GCK DEGWLLSLSGTVSFKNARDLREAAPLIPQRQLLVETDAPFLTPHPYRGAP
Mvan_4802|M.vanbaalenii_PYR-1 DEGWLLSLSGTVSFKNARDLREAVPLIPADQMLVETDAPFLTPHPFRGAP
MSMEG_5440|M.smegmatis_MC2_155 DAGWVLSLSGTVSFKNARDLQEAARLIPQGQMLVETDAPFLTPHPFRGAP
TH_0978|M.thermoresistible__bu EAGWVLSLSGTVSFRNARALHEAAPLIPPGQMLVETDAPYLTPHPYRGAP
Rv1008|M.tuberculosis_H37Rv DAGWLLSLSGTVSFRTARELREAVPLMPVEQLLVETDAPYLTPHPHRGLA
Mb1035|M.bovis_AF2122/97 DAGWLLSLSGTVSFRNARELREAVPLMPVEQLLVETDAPYLTPHPHRGLA
MMAR_4480|M.marinum_M AAGWLLSLSGTVSFRNARELREAVPLIPLDQLLVETDAPFLTPHPYRGAA
MUL_4653|M.ulcerans_Agy99 AAGWLLSLSGTVSFRNARELREAVPLIPLDQLLVETDAPFLTPHPYRGAA
MLBr_00239|M.leprae_Br4923 DAGWMLSLSGTVSFRNARPLREAVPLIPAQQLLVETDAPFLTPHPHRGLA
MAV_1146|M.avium_104 DAGWLLSLSGTVSFRNARALREAVPLIPPGQLLVETDAPFLTPHPHRGTA
MAB_1129|M.abscessus_ATCC_1997 AEGWILSLSGTVSFKNARALQEAAALIPDEQLLVETDAPYLTPHPHRGAP
**:*********:.** *:**. *:* *:*******:*****.** .
Mflv_1931|M.gilvum_PYR-GCK NEPYCLPYTVRALADLLGRPAEELAEETSATAVRTYGLAEAWA-------
Mvan_4802|M.vanbaalenii_PYR-1 NEPYCLPYTVRALAEVLSRPAETIAEQTSATAKRVYGLASCWN-------
MSMEG_5440|M.smegmatis_MC2_155 NEPYCLPYTVRALATLLDRPAEEIAAETAATAERVYRLSESCRF------
TH_0978|M.thermoresistible__bu NEPYCLPYTVRALADLLGRPAEDVAWETADTAVSVYGLDESCRI------
Rv1008|M.tuberculosis_H37Rv NEPYCLPYTVRALAELVNRRPEEVALITTSNARRAYGLGWMRQ-------
Mb1035|M.bovis_AF2122/97 NEPYCLPYTVRALAELVNRRPEEVALITTSNARRAYGLGWMRQ-------
MMAR_4480|M.marinum_M NEPYCLPYTVRALAELLNGSAEELAQVTASNARRVYGLG-----------
MUL_4653|M.ulcerans_Agy99 NEPYCLPYTVRALAELLNGSAEELAQVTARNARRVYGLG-----------
MLBr_00239|M.leprae_Br4923 NEPYCLPYTVRAVAELVERSPEELAQITTSNALRVYKLGSV---------
MAV_1146|M.avium_104 NEPYCLPYTVRAIAELVDRRPEELAAVTTDNARRVYGLA-----------
MAB_1129|M.abscessus_ATCC_1997 NEPYCLPYTVRALAEITGRTADRLAEASSTTAIAVFKIVDELPPRDAFVT
************:* : .: :* :: .* .: :
Mflv_1931|M.gilvum_PYR-GCK --
Mvan_4802|M.vanbaalenii_PYR-1 --
MSMEG_5440|M.smegmatis_MC2_155 --
TH_0978|M.thermoresistible__bu --
Rv1008|M.tuberculosis_H37Rv --
Mb1035|M.bovis_AF2122/97 --
MMAR_4480|M.marinum_M --
MUL_4653|M.ulcerans_Agy99 --
MLBr_00239|M.leprae_Br4923 --
MAV_1146|M.avium_104 --
MAB_1129|M.abscessus_ATCC_1997 VS