For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIGNCSGFYGDRLSAMREMLTGGELDYLTGDYLAELTMLILGRDRMKNPDRGYAKTFLAQLEDCLGLAHD RGVRIVTNAGGLNPAGLANAVRALAARLGIPAQVAHVEGDDLQPRAAELGLGTPLTANAYLGAWGIVDCF ERGADVVVTGRVTDASVVVGAAAAHFGWGRTDYHRLAGAVVAGHVIECGVQATGGNYAFFTEIGDLTHAG FPLAEIAADGSSVITKHHGTGGLVSVDTITAQLLYEITGARYANPDVTARMDSVELSPDGPDRVRISGVI GEPPPPTYKVSLNSIGGFRNAMTFVLTGLDIDAKADLVRRQLEAALTVKPAELQWTLARTDHPDADTEET ASALLTCVARDPDPANVGRQFSSAAVELALASYPGFTATAPPGDGQVYGVFTPGYVDAGKVAHIAVHADG TRTEIPCATETLELAPAHPPALPDPLPAGPTRRVPLGLIAGARSGDKGGSANVGVWVRTDEQWRWLAHTL TVELLKELLPETAGLVVTRHVLPNLRALNFVIEAILGQGVAYQARFDPQAKGLGEWLRSRHVEIPETLL*
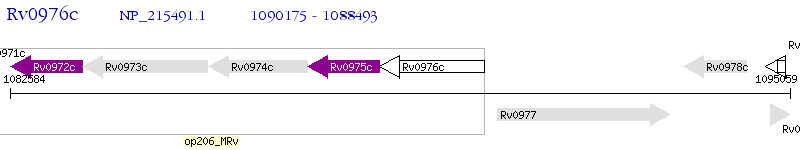
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0976c | - | - | 100% (560) | hypothetical protein Rv0976c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1001c | - | 0.0 | 100.00% (560) | hypothetical protein Mb1001c |
| M. gilvum PYR-GCK | Mflv_1885 | - | 0.0 | 75.89% (564) | hypothetical protein Mflv_1885 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1074c | - | 0.0 | 77.21% (544) | hypothetical protein MAB_1074c |
| M. marinum M | MMAR_4531 | - | 0.0 | 87.68% (560) | hypothetical protein MMAR_4531 |
| M. avium 104 | MAV_1092 | - | 0.0 | 85.89% (560) | hypothetical protein MAV_1092 |
| M. smegmatis MC2 155 | MSMEG_5490 | - | 0.0 | 79.75% (563) | hypothetical protein MSMEG_5490 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4703 | - | 0.0 | 87.32% (560) | hypothetical protein MUL_4703 |
| M. vanbaalenii PYR-1 | Mvan_4846 | - | 0.0 | 79.46% (560) | hypothetical protein Mvan_4846 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0976c|M.tuberculosis_H37Rv -------------MRIGNCSGFYGDRLSAMREMLTGGELDYLTGDYLAEL
Mb1001c|M.bovis_AF2122/97 -------------MRIGNCSGFYGDRLSAMREMLTGGELDYLTGDYLAEL
MMAR_4531|M.marinum_M MASRSDASG---AVRIANCSGFYGDRLSAMREMLTGGEVDYLTGDYLAEL
MUL_4703|M.ulcerans_Agy99 MASRSDASG---AVRIANCSGFYGDRLSAMREMLTGGEVDYLTGDYLAEL
MAV_1092|M.avium_104 MASRPGPPGDPGAVRIANCSGFYGDRLSAMREMLTGGEVDYLTGDYLAEL
MAB_1074c|M.abscessus_ATCC_199 -----------------------------MHEMLTGGELDYLTGDYLAEL
MSMEG_5490|M.smegmatis_MC2_155 MTSG--------SVRIGNCSGFYGDRMAAMREMLTGGELDYLTGDYLAEL
Mflv_1885|M.gilvum_PYR-GCK MTPMGDPVPVRPPVRIANCSGFYGDRLSAMHEMLAGGELDYLTGDYLAEL
Mvan_4846|M.vanbaalenii_PYR-1 MT----TAPFRTPVRIGNCSGFYGDRLAAMREMLTGGELDYLTGDYLAEL
*:***:***:***********
Rv0976c|M.tuberculosis_H37Rv TMLILGRDRMKNPDRGYAKTFLAQLEDCLGLAHDRGVRIVTNAGGLNPAG
Mb1001c|M.bovis_AF2122/97 TMLILGRDRMKNPDRGYAKTFLAQLEDCLGLAHDRGVRIVTNAGGLNPAG
MMAR_4531|M.marinum_M TMLILGRDRMKNPERGYAKTFLAQLEDCLGLAHERGVRIVANAGGLNPAG
MUL_4703|M.ulcerans_Agy99 TMLILGRDRMKNPERGYAKTFLAQLEDCLGLAHDRGVRIVANAGGLNPAG
MAV_1092|M.avium_104 TMLILGRDRMKHPERGYAKTFLTQLEDCLGEARDRGVRIVANAGGLNPAG
MAB_1074c|M.abscessus_ATCC_199 TMLILGRDRAKAPERGYAKTFLRQLEQCLGLAQDKGVRIVANAGGLNPAG
MSMEG_5490|M.smegmatis_MC2_155 TMLILARDRAKDPGRGYARTFLRQLEESLGLALDKGVKIVANAGGLNPAG
Mflv_1885|M.gilvum_PYR-GCK TMLILGRDRMKDPDRGYARTFLRQLEECVGLAADRGVRIVANAGGLNPAA
Mvan_4846|M.vanbaalenii_PYR-1 TMLILGRDRMKNPDRGYARTFLRQLEECLGLAKDRGVRIVANAGGLNPAG
*****.*** * * ****:*** ***:.:* * ::**:**:********.
Rv0976c|M.tuberculosis_H37Rv LANAVRALAARLGIPAQVAHVEGDDLQPRAAELGL---GTPLTANAYLGA
Mb1001c|M.bovis_AF2122/97 LANAVRALAARLGIPAQVAHVEGDDLQPRAAELGL---GTPLTANAYLGA
MMAR_4531|M.marinum_M LADAIRALTERLGIGARIAHVEGDDLAPRAAELGL---GTPLTANAYLGA
MUL_4703|M.ulcerans_Agy99 LADAIRALAECLGIGARIAHVEGDDLAPRAAELGL---GTSLTANAYLGA
MAV_1092|M.avium_104 LADAIRDLAARLRLDVRVAHVEGDDLLARAAELGL---GSPLTANAYLGA
MAB_1074c|M.abscessus_ATCC_199 LADAIRALSEKLGVPTNVAHVEGDDLLPRAAELGL---GSPLTANAYLGA
MSMEG_5490|M.smegmatis_MC2_155 LAEAVRALAGRLGLSVAVAHVEGDDLLPRAAELGFSESGDPLAANAYLGA
Mflv_1885|M.gilvum_PYR-GCK LARAVRSLTEKLGVAVSVAHVEGDDLLSRAEALGL---GDPLTANAYLGA
Mvan_4846|M.vanbaalenii_PYR-1 LAAAVRALADKLGVAITVAHVEGDDLLARAEQLRL---GNPLTANAYLGA
** *:* *: * : :******** .** * : * .*:*******
Rv0976c|M.tuberculosis_H37Rv WGIVDCFERGADVVVTGRVTDASVVVGAAAAHFGWGRTDYHRLAGAVVAG
Mb1001c|M.bovis_AF2122/97 WGIVDCFERGADVVVTGRVTDASVVVGAAAAHFGWGRTDYHRLAGAVVAG
MMAR_4531|M.marinum_M WGIVDCLNSGADVVVTGRVTDASVIVGSAAAHFGWGRTDYNQLAGAVVAG
MUL_4703|M.ulcerans_Agy99 WGIVDCLNSGADVVVTGRVTDASVIVGSAAAHFGWGRTDYNQLAGAVVAG
MAV_1092|M.avium_104 WGIVDCLGDGANVVVTGRVTDASVIVGAAAAHFGWERSDYDRLAGAVVAG
MAB_1074c|M.abscessus_ATCC_199 WGIVECLNSGANVVVTGRVTDASVIVGPAAAHFGWRRDDYDKLAGAVAAG
MSMEG_5490|M.smegmatis_MC2_155 WGIVDCLNAGADVVVTGRVTDASVTVGPAAAHFGWARTDYDALAGAVAAG
Mflv_1885|M.gilvum_PYR-GCK WGIVECLTARADIVVTGRVTDASVVVGPAAAHFGWRRTDFDRIAGAVAAG
Mvan_4846|M.vanbaalenii_PYR-1 WGIVDCLDSGADVVVTGRVTDASVIVGPAASHFGWQPTDHDLIAGAVAAG
****:*: *::*********** **.**:**** *.. :****.**
Rv0976c|M.tuberculosis_H37Rv HVIECGVQATGGNYAFFT----EIGDLTHAGFPLAEIAADGSSVITKHHG
Mb1001c|M.bovis_AF2122/97 HVIECGVQATGGNYAFFT----EIGDLTHAGFPLAEIAADGSSVITKHHG
MMAR_4531|M.marinum_M HVIECGVQATGGNYAFFT----EVPDLTYAGFPLAEVSADGTSVITKHPG
MUL_4703|M.ulcerans_Agy99 HVIECGVQATGGNYAFFT----EVPDLTYAGFPLAEVSADGTSVITKHPG
MAV_1092|M.avium_104 HVIECGVQATGGNYSFFT----EVPDLTHAGFPLAEVHADGSSVITKHPG
MAB_1074c|M.abscessus_ATCC_199 HIIECGTQATGGNYAFFT----EIADLSHAGFPLAEVYEDGSSVITKHAG
MSMEG_5490|M.smegmatis_MC2_155 HVIECGAQATGGNYAFFT----EIEDLLHPGFPIAEIAADGSSVITKHPG
Mflv_1885|M.gilvum_PYR-GCK HVIECGTQATGGNFAFFARDFPDLTALTRPGFPIAEIHADGTSVITKHPG
Mvan_4846|M.vanbaalenii_PYR-1 HVIECGTQASGGNFSFFT----ELADLSHPGFPIAEIHADGTSVITKHPG
*:****.**:***::**: :: * .***:**: **:****** *
Rv0976c|M.tuberculosis_H37Rv TGGLVSVDTITAQLLYEITGARYANPDVTARMDSVELSPDGPDRVRISGV
Mb1001c|M.bovis_AF2122/97 TGGLVSVDTITAQLLYEITGARYANPDVTARMDSVELSPDGPDRVRISGV
MMAR_4531|M.marinum_M TGGLVSPDTVTAQLLYEISGARYANPDVTARMDTIQLSSDGPDRVRISGV
MUL_4703|M.ulcerans_Agy99 TGGLVSPDTVTAQLLYEISGARYANPDVTARMDTIQLSSDGPDRVRIGGV
MAV_1092|M.avium_104 TGGLVSVDTVTAQLLYEITGARYANPDVTARMDTVELSSDGPDRVRISGV
MAB_1074c|M.abscessus_ATCC_199 TGGAVSVGTVTAQLLYEITGGRYANPDVTTRIDSIELSQEGPDRVRVSGV
MSMEG_5490|M.smegmatis_MC2_155 TGGRVDVGTVTAQLLYEIGGARYANPDVTLRVDSLRLDPDGPDRVRISGV
Mflv_1885|M.gilvum_PYR-GCK TGGAVTTGTVTAQLLYEITGARYANPDATLRVDSIELSADGVDRVRISGV
Mvan_4846|M.vanbaalenii_PYR-1 TGGAVTVDTVTAQLLYEIGGARYANPDATLRVDSIELTSQGPDRVRISGV
*** * .*:******** *.******.* *:*::.* :* ****:.**
Rv0976c|M.tuberculosis_H37Rv IGEPPPPTYKVSLNSIGGFRNAMTFVLTGLDIDAKADLVRRQLEAALTVK
Mb1001c|M.bovis_AF2122/97 IGEPPPPTYKVSLNSIGGFRNAMTFVLTGLDIDAKADLVRRQLEAALTVK
MMAR_4531|M.marinum_M IGEAPPPTLKVSLNSIGGFRNAMTFVLTGLNIEAKAELVQRQLEATLTVK
MUL_4703|M.ulcerans_Agy99 IGEAPPQTLKVSLNSIGGFRNAMTFVLTGLNIEAKAELVQRQLEATLTVK
MAV_1092|M.avium_104 RGEPPPPTYKVSLNSIGGFRNSMTFVLTGLDIEAKAELVRRQLQSALAVK
MAB_1074c|M.abscessus_ATCC_199 TGEAPPPDLKVSLNALGGFRNQMTFVLTGLDIDAKADLTRRQLEAALTSP
MSMEG_5490|M.smegmatis_MC2_155 VGEPPPPTYKVSLNRIGGFRNATTFVLTGLDIEAKAQLVRNQLECALKTE
Mflv_1885|M.gilvum_PYR-GCK RGEPPPPTAKVGLNTLGGFRNEMTFVLTGLDVEAKAGLVRHQLEAGLTVR
Mvan_4846|M.vanbaalenii_PYR-1 KGEPPPPTLKVSLNSLGGFRNEVTFVLTGLEIEAKADLVRRQLETSLPVR
**.** **.** :***** *******:::*** *.:.**: *
Rv0976c|M.tuberculosis_H37Rv PAELQWTLARTDHPDADTEETASALLTCVARDPDPANVGRQFSSAAVELA
Mb1001c|M.bovis_AF2122/97 PAELQWTLARTDHPDADTEETASALLTCVARDPDPANVGRQFSSAAVELA
MMAR_4531|M.marinum_M PAELEWTLARTDHVDADTEEAASALLHCVVRDPDPANVGRKFSSAAVELA
MUL_4703|M.ulcerans_Agy99 PAELEWTLARTDHVDADTEEAASALLHCVVRDPDPANVGRKFSSAAVELA
MAV_1092|M.avium_104 PAELQWSLARTDHLDADTEEAASALLHCVVRDPDPANVGRQFSSAAVELA
MAB_1074c|M.abscessus_ATCC_199 PEDIQWTLGRTDHPNADTEQAASALLHVMVRDSDPNKVGRQFSNAAVELA
MSMEG_5490|M.smegmatis_MC2_155 PAELEWTLARTDHPDAGTEEAASALLHCVVRDPDPNTVGRQFSSAAVELA
Mflv_1885|M.gilvum_PYR-GCK PAEMDWTLARTDRPDADSEQAASALLRCVVRDPDPDVVGRRFSSAAVELA
Mvan_4846|M.vanbaalenii_PYR-1 PAELEWTLARTDHPDATTEQEASALLRCVVRDPDPATVGRQFSSAAVELA
* :::*:*.***: :* :*: ***** :.**.** ***:**.******
Rv0976c|M.tuberculosis_H37Rv LASYPGFTATAPPGDGQVYGVFTPGYVDAGKVAHIAVHADGTRTEIPCAT
Mb1001c|M.bovis_AF2122/97 LASYPGFTATAPPGDGQVYGVFTPGYVDAGKVAHIAVHADGTRTEIPCAT
MMAR_4531|M.marinum_M LASYPGFTSTAPPGDGQVYGVFVPGYVDAKLVPHIAVHADGTRTDIPCAT
MUL_4703|M.ulcerans_Agy99 LASYPGFTSTAPPGDGQVYGVFVPGYVDAKLVPHIAVHADGTRTYIPCAT
MAV_1092|M.avium_104 LASYPGFHVTAPPGDGQVYGVFTAGYVDADKVPHIAVHADGTRVDIPCAT
MAB_1074c|M.abscessus_ATCC_199 LAGYPGFHVTTPPKDGEPYGVFTPGYVPAEKVPHVAVLADGTRIDIAQAT
MSMEG_5490|M.smegmatis_MC2_155 LASYPGFTATAPPGDAQIYGIFTPGYVAAAEVAHVAVHADGTRVAIPPTS
Mflv_1885|M.gilvum_PYR-GCK LASYPGFTTTAPPGDGQVYGVFTAAYVPATEVPHTAVRPDGTHVDIEAAP
Mvan_4846|M.vanbaalenii_PYR-1 LASYPGFTSTAPPGDGQVYGVFTAAYVPAHDVAHVAVHSDGVRIDIPAAT
**.**** *:** *.: **:*...** * *.* ** .**.: * :.
Rv0976c|M.tuberculosis_H37Rv ETLELAPAHPPALPDPLPAGPTRRVPLGLIAGARSGDKGGSANVGVWVRT
Mb1001c|M.bovis_AF2122/97 ETLELAPAHPPALPDPLPAGPTRRVPLGLIAGARSGDKGGSANVGVWVRT
MMAR_4531|M.marinum_M ETLELESVDPPALPDPLPTGPTRRVPLGLIAGARSGDKGGSANVGLWVRT
MUL_4703|M.ulcerans_Agy99 ETLELESVDPPALPDPLPTGPTRRVPLGLIAGARSGDKGGSANVGLWVRT
MAV_1092|M.avium_104 DTLELAPVDDPALPEPLPDGPVRRAPLGTIAGARSGDKGGSANVGVWVRT
MAB_1074c|M.abscessus_ATCC_199 QTQPLAEAPAAALPEPLPAGPVRRAPLGILAGARSGDKGGSANVGVWVRT
MSMEG_5490|M.smegmatis_MC2_155 ETRELGPVPEPAVAPPDP-GPTVRAPLGLIAGARSGDKGGSAYVGVWVRS
Mflv_1885|M.gilvum_PYR-GCK QTLALADLPAPVLPDAPTPGPTRRVPLGTIAGARSGDKGGSANVGVWVSS
Mvan_4846|M.vanbaalenii_PYR-1 DTEELTDPPQPQLPPQREFGVTRRVPLGLIAGARSGDKGGSANVGVWVRT
:* * . :. * . *.*** :************ **:** :
Rv0976c|M.tuberculosis_H37Rv DEQWRWLAHTLTVELLKELLPETAGLVVTRHVLPNLRALNFVIEAILGQG
Mb1001c|M.bovis_AF2122/97 DEQWRWLAHTLTVELLKELLPETAGLVVTRHVLPNLRALNFVIEAILGQG
MMAR_4531|M.marinum_M DEQWRWLANTLTVELLKELLPEAADLQVTRYALPNLRALNFVIEGILGQG
MUL_4703|M.ulcerans_Agy99 DEQWRWLANTLTVELLKELLPEAANLQVTRYALPNLRALNFVIEGILGQG
MAV_1092|M.avium_104 DEQWRWLAHTLTVERLTELLPEAAEFAVTRHLLPNLRAVNFVIDGILGQG
MAB_1074c|M.abscessus_ATCC_199 DEEYRWLAHALTVDALRELLPETEHLEVARYLLPKLRAVNFVIQDILGKG
MSMEG_5490|M.smegmatis_MC2_155 DASYRWLVHTLTVDVLRDLLPEAAELPVTRHVLPNLRALNFVIDDILGKG
Mflv_1885|M.gilvum_PYR-GCK AQQWAWLVHTLTVDTVRELLPEVADLPVTRHVLPNLRAVNFVIDGILGKG
Mvan_4846|M.vanbaalenii_PYR-1 DDQWTWLVHTLTVEKVRELLPEAADLPVTRHVLPNLRAVNFVIDGILGRG
.: **.::***: : :****. : *:*: **:***:****: ***:*
Rv0976c|M.tuberculosis_H37Rv VAYQARFDPQAKGLGEWLRSRHVEIPETLL---
Mb1001c|M.bovis_AF2122/97 VAYQARFDPQAKGLGEWLRSRHVEIPETLL---
MMAR_4531|M.marinum_M VAYQARFDPQAKGLGEWLRSRHLDIPESLL---
MUL_4703|M.ulcerans_Agy99 VAYQARFDPQAKGLGEWLRSRHLDIPESLL---
MAV_1092|M.avium_104 VAYQARFDPQAKGLGEWLRGRYLDIPESLL---
MAB_1074c|M.abscessus_ATCC_199 VAYQARFDPQAKGIGEWLRSREIDIPESLLEGK
MSMEG_5490|M.smegmatis_MC2_155 VACQARFDPQAKGLGEWLRSRHLDIPKDLLP--
Mflv_1885|M.gilvum_PYR-GCK VAYQARFDPQAKGLGEWLRSRHIDIPEELLA--
Mvan_4846|M.vanbaalenii_PYR-1 VAYQARFDPQAKGLGEWLRSRCIEIPEELLP--
** **********:*****.* ::**: **