For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGILDKVKNLLSQNADKVETVINKAGEFVDEQTQGNYSDAIHKLHDAASNVVGMSDQQS
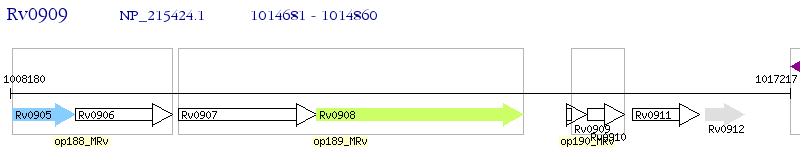
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0909 | - | - | 100% (59) | hypothetical protein Rv0909 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0933 | - | 5e-28 | 98.31% (59) | hypothetical protein Mb0933 |
| M. gilvum PYR-GCK | Mflv_1759 | - | 2e-13 | 55.17% (58) | hypothetical protein Mflv_1759 |
| M. leprae Br4923 | MLBr_02114 | - | 6e-12 | 61.90% (42) | hypothetical protein MLBr_02114 |
| M. abscessus ATCC 19977 | MAB_0964 | - | 7e-09 | 43.14% (51) | hypothetical protein MAB_0964 |
| M. marinum M | MMAR_4620 | - | 1e-17 | 71.70% (53) | hypothetical protein MMAR_4620 |
| M. avium 104 | MAV_1032 | - | 6e-16 | 62.50% (56) | hypothetical protein MAV_1032 |
| M. smegmatis MC2 155 | MSMEG_5635 | - | 3e-15 | 58.62% (58) | hypothetical protein MSMEG_5635 |
| M. thermoresistible (build 8) | TH_4122 | - | 2e-11 | 51.92% (52) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0161 | - | 9e-18 | 71.70% (53) | hypothetical protein MUL_0161 |
| M. vanbaalenii PYR-1 | Mvan_4988 | - | 4e-13 | 57.69% (52) | hypothetical protein Mvan_4988 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1759|M.gilvum_PYR-GCK ----MGFLDKAKDLLSKNADKVDTVIDKAGNLVDKKTQGKYSSTVDKVQE
Mvan_4988|M.vanbaalenii_PYR-1 ----MGFLDKAKDLLSKNADKVDTVIDKAGDLVDKKTQGKYASTVDKVQE
MSMEG_5635|M.smegmatis_MC2_155 ----MGFLDKAKDLLSQNADKVDTVIDKAGDMIDEKTQGKYAQHVDKAQD
Rv0909|M.tuberculosis_H37Rv ----MGILDKVKNLLSQNADKVETVINKAGEFVDEQTQGNYSDAIHKLHD
Mb0933|M.bovis_AF2122/97 ----MGILDKVKNLLSQNADKVETVINKAGEFVDEQTQGNYSDAIHKLQD
MMAR_4620|M.marinum_M ----MGILDKAKDLLAQNADKVETVIDKAGEFVDEKTEGKYSDTIHKVQE
MUL_0161|M.ulcerans_Agy99 ----MGILDKAKDLLAQNADKVETVIDKAGEFVDEKTEGKYSDTIHKVQE
MLBr_02114|M.leprae_Br4923 ------MLDKAKDILVQNADKVETVLDKAGEFVDEKAKGKYTDTIYQVAE
MAV_1032|M.avium_104 ----MGFLDKAKDLLSQNADKVEMAIDKAGEFVDEKTQGKYSDTIHQVQE
TH_4122|M.thermoresistible__bu ----MGFLDKVKDWLGKNPDKADAAIEKAGDFVDQRTQGRYAEHVDKVQD
MAB_0964|M.abscessus_ATCC_1997 MADFKGLIDKLKSLLAGNKDKVNEAVDKVGDTIDSKTGGKYSAVVDKVQD
::** *. * * **.: .::*.*: :*.:: *.*: : : :
Mflv_1759|M.gilvum_PYR-GCK AARKAVADNT----DQGRVPPTQGSGQVPPSSPPPSSPPPPATPPPPAPP
Mvan_4988|M.vanbaalenii_PYR-1 AAKKAVADN------AGPVPPGQQSGHVPPPQPPPT------TPPQQQPP
MSMEG_5635|M.smegmatis_MC2_155 AAKNAIHKD-------DPQEPQQ---------------------------
Rv0909|M.tuberculosis_H37Rv AASNVVG------------MSDQQS-------------------------
Mb0933|M.bovis_AF2122/97 AASNVVG------------MSDQQS-------------------------
MMAR_4620|M.marinum_M GAKKVAG------------PGAAGEQQN----------------------
MUL_0161|M.ulcerans_Agy99 GAKKVAG------------PGAAGEQQN----------------------
MLBr_02114|M.leprae_Br4923 ETKKAAS------------FDTFG--------------------------
MAV_1032|M.avium_104 QAKKATG------------ATETGDQQG----------------------
TH_4122|M.thermoresistible__bu AARKYVTGGQAQQGNPTPPHPDQGDQGNPTPQQP----------------
MAB_0964|M.abscessus_ATCC_1997 AAKSAVEKVEG------AAAPKDGDAQ-----------------------
: .
Mflv_1759|M.gilvum_PYR-GCK TAPPTS
Mvan_4988|M.vanbaalenii_PYR-1 TAPPTS
MSMEG_5635|M.smegmatis_MC2_155 ------
Rv0909|M.tuberculosis_H37Rv ------
Mb0933|M.bovis_AF2122/97 ------
MMAR_4620|M.marinum_M ------
MUL_0161|M.ulcerans_Agy99 ------
MLBr_02114|M.leprae_Br4923 ------
MAV_1032|M.avium_104 ------
TH_4122|M.thermoresistible__bu ------
MAB_0964|M.abscessus_ATCC_1997 ------