For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRRAIHSGRAAPRRSGNSHLVLRNRVPSSKDSPRRRPHHEFMTESIGEPLSTNLIERYLRARGRRYFRGH HDAEFFFVANAHLRLHVHLEISPAYRDVFTIRVSPAYFFPATDHTRLAEIVNAWNLQNHEVTAIVHGSSD PHRIGVAAERSLIRDRIRFDDFATFVDNAVSAATELFGQLTAAGLPPTATPPLLRDAG*
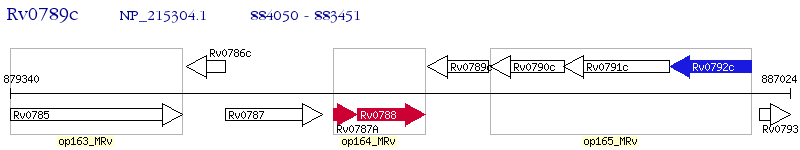
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0789c | - | - | 100% (199) | hypothetical protein Rv0789c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0813c | - | 1e-114 | 100.00% (199) | hypothetical protein Mb0813c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4896 | - | 9e-45 | 61.69% (154) | hypothetical protein MMAR_4896 |
| M. avium 104 | MAV_0737 | - | 3e-43 | 57.89% (152) | hypothetical protein MAV_0737 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0402 | - | 3e-44 | 61.04% (154) | hypothetical protein MUL_0402 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4896|M.marinum_M ----------------------MT------------------------EP
MUL_0402|M.ulcerans_Agy99 ----------------------MT------------------------EP
MAV_0737|M.avium_104 -------MAPSVRKFAGGSMFVMTN-----------------------EQ
Rv0789c|M.tuberculosis_H37Rv MSRRAIHSGRAAPRRSGNSHLVLRNRVPSSKDSPRRRPHHEFMTESIGEP
Mb0813c|M.bovis_AF2122/97 MSRRAIHSGRAAPRRSGNSHLVLRNRVPSSKDSPRRRPHHEFMTESIGEP
: *
MMAR_4896|M.marinum_M LCATLIERYLCTRGRRYFRGQHDGEFFFVTDAHPQRLHVHLEISPAHPEV
MUL_0402|M.ulcerans_Agy99 LCATLIERYLCTRGRRYFRGQHDGEFFFVTDAHPQRLHVHLEISPAHPEV
MAV_0737|M.avium_104 SCANLVERYLRTRGRRYFRGHHDGEYFFVT-AAPRRLHVHFEISPAHDDV
Rv0789c|M.tuberculosis_H37Rv LSTNLIERYLRARGRRYFRGHHDAEFFFVANAHLR-LHVHLEISPAYRDV
Mb0813c|M.bovis_AF2122/97 LSTNLIERYLRARGRRYFRGHHDAEFFFVANAHLR-LHVHLEISPAYRDV
.:.*:**** :********:**.*:***: * : ****:*****: :*
MMAR_4896|M.marinum_M FAIRVAPACFFPAGDRALLARFADRWNTQRRGVTAIVHNSSDPQRIGVTA
MUL_0402|M.ulcerans_Agy99 FAIRVAPACFFPAGDRALLARFADRWNTQRRGVTAIVHNSSDPQRIGVTA
MAV_0737|M.avium_104 LIIRVSPGCYFPASDRPWLTHFSDRWNRLGRRVTAIVHGSSDPQRIGVTA
Rv0789c|M.tuberculosis_H37Rv FTIRVSPAYFFPATDHTRLAEIVNAWNLQNHEVTAIVHGSSDPHRIGVAA
Mb0813c|M.bovis_AF2122/97 FTIRVSPAYFFPATDHTRLAEIVNAWNLQNHEVTAIVHGSSDPHRIGVAA
: ***:*. :*** *:. *:.: : ** : ******.****:****:*
MMAR_4896|M.marinum_M YKSQWIAGPVDFSEFAVFVDRAIGASIELFGELNRVAELP-QTPPLMRDA
MUL_0402|M.ulcerans_Agy99 YKSQWIAGPVDFSEFAVFVDRAIGASIELFGELNRVAELP-QTSPLMRDA
MAV_0737|M.avium_104 RKSQWIRNGVGFTDFAAFLDETIADATQLFEELN-PAAPP-AHAPLLRDA
Rv0789c|M.tuberculosis_H37Rv ERS-LIRDRIRFDDFATFVDNAVSAATELFGQLTAAGLPPTATPPLLRDA
Mb0813c|M.bovis_AF2122/97 ERS-LIRDRIRFDDFATFVDNAVSAATELFGQLTAAGLPPTATPPLLRDA
:* * . : * :**.*:*.::. : :** :*. . * .**:***
MMAR_4896|M.marinum_M G
MUL_0402|M.ulcerans_Agy99 G
MAV_0737|M.avium_104 G
Rv0789c|M.tuberculosis_H37Rv G
Mb0813c|M.bovis_AF2122/97 G
*