For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHGARTGVSFYAYAMTDHDQTAARREIADALLAALERRHEVADAIVEAANKAAAVEAIVNLLGTSHLAAE AVMSMSFDQLTQDARTKIIAELDDLNKQLSFTVKERPASSGEGLELRPFSPDEDRDIFARRTEEMGAAGD GSGGPAGSVDDEIRAAQKRVDDEEAAWFVAVDSGVKVGMVFGELVHGEVDVRIWIHPDHRKKGYGTAALR KSRSEMAWAFPAVPMVARAPAAQPAQPGSAGR
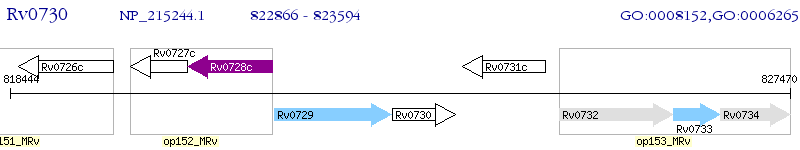
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0730 | - | - | 100% (242) | hypothetical protein Rv0730 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0751 | - | 1e-136 | 100.00% (242) | hypothetical protein Mb0751 |
| M. gilvum PYR-GCK | Mflv_4079 | - | 1e-67 | 59.82% (219) | hypothetical protein Mflv_4079 |
| M. leprae Br4923 | MLBr_01835 | - | 1e-101 | 83.49% (218) | hypothetical protein MLBr_01835 |
| M. abscessus ATCC 19977 | MAB_2473c | - | 3e-85 | 71.76% (216) | hypothetical protein MAB_2473c |
| M. marinum M | MMAR_1067 | - | 1e-110 | 90.50% (221) | hypothetical protein MMAR_1067 |
| M. avium 104 | MAV_4436 | - | 1e-100 | 81.45% (221) | hypothetical protein MAV_4436 |
| M. smegmatis MC2 155 | MSMEG_6565 | - | 4e-85 | 70.00% (220) | hypothetical protein MSMEG_6565 |
| M. thermoresistible (build 8) | TH_1397 | - | 2e-77 | 64.84% (219) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0825 | - | 1e-113 | 89.87% (227) | hypothetical protein MUL_0825 |
| M. vanbaalenii PYR-1 | Mvan_2264 | - | 2e-65 | 58.26% (230) | hypothetical protein Mvan_2264 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0730|M.tuberculosis_H37Rv -------MHGARTGVSFYAYAMTDHDQTAAR-------------------
Mb0751|M.bovis_AF2122/97 -------MHGARTGVSFYAYAMTDHDQTAAR-------------------
MMAR_1067|M.marinum_M ---------------------MTDHDHTAAR-------------------
MUL_0825|M.ulcerans_Agy99 MGRPDGYAGSAFPTASFYAYAMTDHDHTAAR-------------------
MLBr_01835|M.leprae_Br4923 ---------------------MTDHDQTAAR-------------------
MAV_4436|M.avium_104 ---------------------MTDQDRKAAR-------------------
MAB_2473c|M.abscessus_ATCC_199 ---------------------MTDHDRLATR-------------------
MSMEG_6565|M.smegmatis_MC2_155 --------------MPATLGGMTDPDKAAAR-------------------
Mflv_4079|M.gilvum_PYR-GCK ---------------------MTEHDSTATR-------------------
Mvan_2264|M.vanbaalenii_PYR-1 ---------------------MTDPDATAARMTTREMATAASREMATAAS
TH_1397|M.thermoresistible__bu ---------------------VTDQDQRVDR-------------------
:*: * . *
Rv0730|M.tuberculosis_H37Rv REIADALLAALERRHEVADAIVEAANKAAAVEAIVNLLGTSHLAAEAVMS
Mb0751|M.bovis_AF2122/97 REIADALLAALERRHEVADAIVEAANKAAAVEAIVNLLGTSHLAAEAVMS
MMAR_1067|M.marinum_M REIADALLAALERRHEVADAIVEAENKAAAVEAIVTLLGTSHLAAEAVMS
MUL_0825|M.ulcerans_Agy99 REIADALLAALERRHEVADAIVEAENKAAAVEAIVTLLATSHLAAEAVMS
MLBr_01835|M.leprae_Br4923 RETADALLKALERRHEIADAVVEAENKPAAVEAIVKLLDTSRLAAEAVMS
MAV_4436|M.avium_104 REIADALLKALERRHEIADVVVESENKAAAVEAIVRLLDTSHLAAEAVMG
MAB_2473c|M.abscessus_ATCC_199 REIADALTSAFERRHEVLDLIVEADDRKSAIDAIATLLGTSHLGGEAVMG
MSMEG_6565|M.smegmatis_MC2_155 RDITDALLSALNRRHEVLDVIVDAEDRNAAIDNVANLLGTSRTAAEAVIG
Mflv_4079|M.gilvum_PYR-GCK REIADAMMKALDRRHELLDAVVASEDYDAAIEAIADLLDASAEASEAVLR
Mvan_2264|M.vanbaalenii_PYR-1 REMATALVHALERRHEVLDAVVASDDYDAAIESLAELLDTSAEAAEAVLR
TH_1397|M.thermoresistible__bu REIADALVRALDRRHEILDAIVDSDDTASALGKIRAMLDTSQAAAEAVLR
*: : *: *::****: * :* : : :*: : :* :* ..***:
Rv0730|M.tuberculosis_H37Rv MSFDQLTQDARTKIIAELDDLNK-QLSFTVKERPASSGEGLELRPFSPDE
Mb0751|M.bovis_AF2122/97 MSFDQLTQDARTKIIAELDDLNK-QLSFTVKERPASSGEGLELRPFSPDE
MMAR_1067|M.marinum_M MSFEQLTQDARKKIIAELDDLNK-QLSFTLTERPASSGESLELRPFSAAE
MUL_0825|M.ulcerans_Agy99 MSFEQLTQDARKKIIAELDDLNK-QLSFTLTERPASSGESLELRPFSAAE
MLBr_01835|M.leprae_Br4923 MSFDRLTKDSRKKILAELEDLNK-QLSFTLQERPASLGESLELRPFSGED
MAV_4436|M.avium_104 MSFDQLTIDSRRKILAELEDLNK-QLSFTVGERPASSGETLELRPFSATE
MAB_2473c|M.abscessus_ATCC_199 MSFDKLTKDERRKNAAELEDLNS-QLTFTLMERPASSGDTLELRPFSGSS
MSMEG_6565|M.smegmatis_MC2_155 LSFAQITKESRRQIAAELDDLNS-QLSFTLQERPASAGDTLELRPFSATD
Mflv_4079|M.gilvum_PYR-GCK LSFDRLTKVSRRRIAAELENLNSKVLTFASSEQTPGSARRLMLRPFSAAD
Mvan_2264|M.vanbaalenii_PYR-1 LSFDRLTKVSRRRIAAELEDLNSRAQNYTPPEQTPGSARRLLLRPFAADQ
TH_1397|M.thermoresistible__bu LSFDRLTRESRRQIARELDDLNS-QLSFVLGERPDSSGDNLALRPFRPDR
:** ::* * : **::**. .:. *:. . . * ****
Rv0730|M.tuberculosis_H37Rv DRDIFARRTEEMGAAGDGSGGPAGSVDDEIRAAQKRVDDEEAAWFVAVDS
Mb0751|M.bovis_AF2122/97 DRDIFARRTEEMGAAGDGSGGPAGSVDDEIRAAQKRVDDEEAAWFVAVDS
MMAR_1067|M.marinum_M DRDIFAARTEEMGAAGDGSGGPAGTVDDEIKAALARVDDEEAAWFVAIDA
MUL_0825|M.ulcerans_Agy99 DRDIFAARTEEMGAAGDGSGGPAGTVDDEIKAALARVDDEEAAWFVAIDA
MLBr_01835|M.leprae_Br4923 DRDIFAARTAEINSAGDGSGGPAGSIDDEIRAALVRVADEEAAWFVAVDS
MAV_4436|M.avium_104 DRDIFAARTQDMGSAGDGSGGPAGDLDDEIRAATARLDDEEAAWFVAVDS
MAB_2473c|M.abscessus_ATCC_199 DADIFAVRTEEVGQAGDGSGAPAGVLEDELQAALARIDDEEAAWFVAVEG
MSMEG_6565|M.smegmatis_MC2_155 DRDIFAARTSDVGASGDGSGAPAADLDEEIRAALRRVDAEEAAWFVAVEG
Mflv_4079|M.gilvum_PYR-GCK DRDVFAERTADVRSAGDGTAAPAGDLGDEIRAAVTRMAAEEAVWLVAEVG
Mvan_2264|M.vanbaalenii_PYR-1 DRDIFAARTEDMRSAGDGTAAPAGDLGDEIAAAVARVAAEEAAWLVAEVG
TH_1397|M.thermoresistible__bu DRELFAARTSDVGAAGDGSGAPAGDLDDEIRAANQRMEAEEAVWLVAEVG
* ::** ** :: :***:..**. : :*: ** *: ***.*:** .
Rv0730|M.tuberculosis_H37Rv GVKVGMVFGELVHGEVDVRIWIHPDHRKKGYGTAALRKSRSEMAWAFPAV
Mb0751|M.bovis_AF2122/97 GVKVGMVFGELVHGEVDVRIWIHPDHRKKGYGTAALRKSRSEMAWAFPAV
MMAR_1067|M.marinum_M GEKVGMVFGELVHGEVDVRIWIHPEHRKKGYGTAALRKSRSEMAWAFPAV
MUL_0825|M.ulcerans_Agy99 GEKVGMVFGELVHGEVDVRIWIHPEHRKKGYGTAALRKSRSEMAWAFPAV
MLBr_01835|M.leprae_Br4923 GAKVGMVFGELVHGEVDVRVWICPDHRKKGYGTAALRKSRSEMAYCFPAV
MAV_4436|M.avium_104 GQKVGMVFGELLNGEVNVRIWIHPDFRKRGYGTAALRKSRTEMAWCFPAV
MAB_2473c|M.abscessus_ATCC_199 PSKVGIVFGELVDGEVNVRIWVHPDHRKKGYGTAALRKSRSVMAAYFPAV
MSMEG_6565|M.smegmatis_MC2_155 GQKVGMVFGELVSGEVNVRIWIHPDFRKRGYGTAALRKSRSEMASYFPAV
Mflv_4079|M.gilvum_PYR-GCK STKVGMVFGELADGEVDVRIWIHPAHRKHGFGTAALRASRSEMAAYFPAV
Mvan_2264|M.vanbaalenii_PYR-1 SAKVGMVFGDLADGEVGVRIWIHPSHRKQGYGTAALRASRSEMAAYFPAV
TH_1397|M.thermoresistible__bu SQPVGMVFGELTSGEVNVRIWIHPEHRKKGYGTAALRKSRSEMAAYFPAV
**:***:* ***.**:*: * .**:*:****** **: ** ****
Rv0730|M.tuberculosis_H37Rv PMVARAPAAQPAQPGSAGR
Mb0751|M.bovis_AF2122/97 PMVARAPAAQPAQPGSAGR
MMAR_1067|M.marinum_M PMVVRAPAANPA-------
MUL_0825|M.ulcerans_Agy99 PMVVRIPAANPA-------
MLBr_01835|M.leprae_Br4923 PMVARAPAADLHSRITAD-
MAV_4436|M.avium_104 PLVVRAPAAKPA-------
MAB_2473c|M.abscessus_ATCC_199 PMVVRAPGVDAP-------
MSMEG_6565|M.smegmatis_MC2_155 PMVVRAPGAVPQR------
Mflv_4079|M.gilvum_PYR-GCK PLVVRAPAAG---------
Mvan_2264|M.vanbaalenii_PYR-1 PLVVRAPAAGA--------
TH_1397|M.thermoresistible__bu PLVVRAPAARG--------
*:*.* *..