For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAA GDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVK
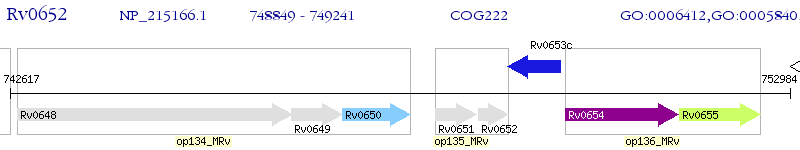
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0652 | rplL | - | 100% (130) | 50S ribosomal protein L7/L12 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0671 | rplL | 9e-66 | 100.00% (130) | 50S ribosomal protein L7/L12 |
| M. gilvum PYR-GCK | Mflv_5100 | rplL | 7e-58 | 88.46% (130) | 50S ribosomal protein L7/L12 |
| M. leprae Br4923 | MLBr_01895 | rplL | 7e-58 | 88.46% (130) | 50S ribosomal protein L7/L12 |
| M. abscessus ATCC 19977 | MAB_3876c | rplL | 8e-57 | 89.23% (130) | 50S ribosomal protein L7/L12 |
| M. marinum M | MMAR_0991 | rplL | 1e-62 | 94.62% (130) | 50S ribosomal protein L7/L12, RplL |
| M. avium 104 | MAV_4507 | rplL | 2e-61 | 93.08% (130) | 50S ribosomal protein L7/L12 |
| M. smegmatis MC2 155 | MSMEG_1365 | rplL | 4e-58 | 89.23% (130) | 50S ribosomal protein L7/L12 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0742 | rplL | 5e-61 | 92.31% (130) | 50S ribosomal protein L7/L12 |
| M. vanbaalenii PYR-1 | Mvan_1253 | rplL | 6e-58 | 87.69% (130) | 50S ribosomal protein L7/L12 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0652|M.tuberculosis_H37Rv MAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPA
Mb0671|M.bovis_AF2122/97 MAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPA
MMAR_0991|M.marinum_M MSKLSTDELLDAFKEMTLLELSDFVKKFEDTFEVTAAAPVAVAAAGAAPA
MUL_0742|M.ulcerans_Agy99 MSKLSTDELLEAFKGMTLLELSDFVKKFEDTFEVTAAAPVAVVAAGAAPA
Mflv_5100|M.gilvum_PYR-GCK MAKLSTDELLDAFKEMTLLELSEFVKQFEETFDVTAAAPVAVAAAGPAAG
Mvan_1253|M.vanbaalenii_PYR-1 MAKLSTDDLLDAFKEMTLLELSEFVKQFEETFDVTAAAPVAVAAAGPAAG
MSMEG_1365|M.smegmatis_MC2_155 MAKLSTEELLDAFKEMTLLELSEFVKQFEETFEVTAAAPVAVAAAPGAAA
MAB_3876c|M.abscessus_ATCC_199 MAKLSTEELLDAFAELTLLELSEFVKAFEEKFEVTAAAPVAVAAVGGAAP
MAV_4507|M.avium_104 MAKMSTDDLLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGPAAG
MLBr_01895|M.leprae_Br4923 MSKLSSDELLDVFKEMTLLELSDFVKKFEETFEVTAAAPVSVAVAGAPAA
*:*:*:::**:.* :******:*** **:.*:*******:*... ..
Rv0652|M.tuberculosis_H37Rv GAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
Mb0671|M.bovis_AF2122/97 GAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
MMAR_0991|M.marinum_M GGAAEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
MUL_0742|M.ulcerans_Agy99 GGAAEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
Mflv_5100|M.gilvum_PYR-GCK GGAAEAAEEQSEFDVILEGAGEKKIGVIKVVREIVSGLGLKEAKDLVDSA
Mvan_1253|M.vanbaalenii_PYR-1 GAAAEAAEEQSEFDVVLESAGEKKIGVIKVVREIVSGLGLKEAKDLVDSA
MSMEG_1365|M.smegmatis_MC2_155 GGAAEAAEEQSEFDVILEGAGEKKIGVIKVVREIVSGLGLKEAKDLVDSA
MAB_3876c|M.abscessus_ATCC_199 -AAADAAEEQSEFDVILESAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
MAV_4507|M.avium_104 GAPAEAAEEQSEFDVILESAGDKKIGVIKVVREIVSGLGLKEAKDLVDGA
MLBr_01895|M.leprae_Br4923 GEAGEAAEEQSEFDVILESAGDKKIGVIKVVREIVSGLGLKEAKDLVDGV
. :**********:**.**:**************************..
Rv0652|M.tuberculosis_H37Rv PKPLLEKVAKEAADEAKAKLEAAGATVTVK
Mb0671|M.bovis_AF2122/97 PKPLLEKVAKEAADEAKAKLEAAGATVTVK
MMAR_0991|M.marinum_M PKPLLEKVAKEAAEEAKAKLEAAGASVSVK
MUL_0742|M.ulcerans_Agy99 PKPLLEKVAKEAAEEAKAKLEAAGASVSVK
Mflv_5100|M.gilvum_PYR-GCK PKPLLEKVNKEAAEDAKAKLEAAGASVTVK
Mvan_1253|M.vanbaalenii_PYR-1 PKPLLEKVNKEAAEDAKGKLEAAGATVTVK
MSMEG_1365|M.smegmatis_MC2_155 PKPLLEKVTKEAADDAKAKLEAAGASVTVK
MAB_3876c|M.abscessus_ATCC_199 PKPLLEKVAKEAADDAKAKLEAAGATVTVK
MAV_4507|M.avium_104 PKPLLEKVAKEAADDAKAKLEAAGATVTVK
MLBr_01895|M.leprae_Br4923 PKLLLEKVAKEAADDAKAKLEATGATVSVK
** ***** ****::**.****:**:*:**