For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TKRTITPMTSMGDLLGPEPILLPGDSDAEAELLANESPSIVAAAHPSASVAWAVLAEGALADDKTVTAYA YARTGYHRGLDQLRRHGWKGFGPVPYSHQPNRGFLRCVAALARAAAAIGETDEYGRCLDLLDDCDPAARP ALGL*
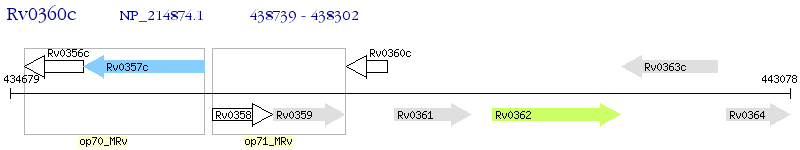
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0360c | - | - | 100% (145) | hypothetical protein Rv0360c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0367c | - | 2e-82 | 100.00% (145) | hypothetical protein Mb0367c |
| M. gilvum PYR-GCK | Mflv_0232 | - | 9e-62 | 81.02% (137) | hypothetical protein Mflv_0232 |
| M. leprae Br4923 | MLBr_00284 | - | 1e-65 | 85.40% (137) | hypothetical protein MLBr_00284 |
| M. abscessus ATCC 19977 | MAB_4252c | - | 2e-56 | 75.74% (136) | hypothetical protein MAB_4252c |
| M. marinum M | MMAR_0672 | - | 2e-64 | 81.29% (139) | hypothetical protein MMAR_0672 |
| M. avium 104 | MAV_4783 | - | 3e-66 | 84.67% (137) | hypothetical protein MAV_4783 |
| M. smegmatis MC2 155 | MSMEG_0754 | - | 1e-60 | 80.60% (134) | hypothetical protein MSMEG_0754 |
| M. thermoresistible (build 8) | TH_1642 | - | 1e-64 | 83.58% (134) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2841 | - | 1e-64 | 81.29% (139) | hypothetical protein MUL_2841 |
| M. vanbaalenii PYR-1 | Mvan_0672 | - | 3e-60 | 78.10% (137) | hypothetical protein Mvan_0672 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0360c|M.tuberculosis_H37Rv MTKRTITPMTSMGDLLGPEPILLPGDSDAEAELLANESPSIVAAAHPSAS
Mb0367c|M.bovis_AF2122/97 MTKRTITPMTSMGDLLGPEPILLPGDSDAEAELLANESPSIVAAAHPSAS
MLBr_00284|M.leprae_Br4923 --------MTSMGDLLGPDPILLPDDSAAEVELRANKDPGTVAAAHPSAS
MMAR_0672|M.marinum_M -----MTPMTSMGDLLGPDPILLPPDSEAEQKLLADESPATVAAAHPSAS
MUL_2841|M.ulcerans_Agy99 -----MTPMTSMGDLLGPDPILLPPDSEAEQKLLADESPATVAAAHPSAS
MAV_4783|M.avium_104 --------MTPMGDLLGPDPILLPGDADAEVELTAGENPGIVAAAHPTAS
Mflv_0232|M.gilvum_PYR-GCK -----MGHMTRMGDLLGPEPVLLPGDPAAEAELDAAENPSIVAAAHPAAS
Mvan_0672|M.vanbaalenii_PYR-1 --------MTRMGDLLGPEPVFLPGDPAAEAELDASENPAIVAAAHPAAS
MSMEG_0754|M.smegmatis_MC2_155 -----------MGDLLGPDPVYLPGDPAAEEELAAGEKAAVVAAAHPSAS
TH_1642|M.thermoresistible__bu -----------MGDLLGPDPVLLPGDSAAEAELAAGENPAIVAAAHPAAS
MAB_4252c|M.abscessus_ATCC_199 -----------MHDLLGPEPVLLPGDDDAESALLNGQDPAAVAAAYPAAS
* *****:*: ** * ** * :... ****:*:**
Rv0360c|M.tuberculosis_H37Rv VAWAVLAEGALADD----KTVTAYAYARTGYHRGLDQLRRHGWKGFGPVP
Mb0367c|M.bovis_AF2122/97 VAWAVLAEGALADD----KTVTAYAYARTGYHRGLDQLRRHGWKGFGPVP
MLBr_00284|M.leprae_Br4923 VAWAALAEGALADD----KATTAYAYARTGYHRGLDQLRCNGWKGFGPVP
MMAR_0672|M.marinum_M VAWAALAEEALDDD----KVITAYAYARTGYHRGLDKLRRNGWKGFGPVP
MUL_2841|M.ulcerans_Agy99 VAWAALAEEALDDD----KVITAYAYARTGYHRGLDKLRRNGWKGFGPVP
MAV_4783|M.avium_104 VAWAALAEQALADD----RAITAYAYARTGYHRGLDQLRRNGWKGFGPVP
Mflv_0232|M.gilvum_PYR-GCK IAWAVLAEQALEAD----QAVAAYAYARTGYHRGLDSLRRNGWKGFGPVP
Mvan_0672|M.vanbaalenii_PYR-1 IAWAVLAEQALDAE----QAVTAYAYARTGYHRGLDALRRNGWKGFGPVP
MSMEG_0754|M.smegmatis_MC2_155 VAWATLAEQALDDD----KAVTAYAYARTGYHRGLDQLRRNGWKGFGPVP
TH_1642|M.thermoresistible__bu IAWAALAEQALADE----QAITAYAYARTGYHRGLDQLRRNGWKGFGPVP
MAB_4252c|M.abscessus_ATCC_199 IAWAELAENALDDDGSIASIVTAYAFARTGYHRGLDQLRRNGWKGFGPVP
:*** *** ** : :***:********** ** :*********
Rv0360c|M.tuberculosis_H37Rv YSHQPNRGFLRCVAALARAAAAIGETDEYGRCLDLLDDCDPAARPALGL-
Mb0367c|M.bovis_AF2122/97 YSHQPNRGFLRCVAALARAAAAIGETDEYGRCLDLLDDCDPAARPALGL-
MLBr_00284|M.leprae_Br4923 YSHEPNRGFLRCVAALARAANAIGETDEYRRCLNLLDDCDPAARNELGL-
MMAR_0672|M.marinum_M YAHESNQGFLRCVAALARAADSIGETDEYRRCLDLLDDCDPTARSVLGF-
MUL_2841|M.ulcerans_Agy99 YAHESNQGFLRCVAALARAADSIGETDEYRRCLDLLDDCDPTARSVLGF-
MAV_4783|M.avium_104 YSHEPNRGFLRCVAALARAADTIGETDEYLRCLDLLDDCDPAARKELGL-
Mflv_0232|M.gilvum_PYR-GCK FAHEPNQGFLRCVAALARAADAIGETPEYQRCLDLLDDCDPAARGRLGLG
Mvan_0672|M.vanbaalenii_PYR-1 FAHEPNQGFLRCVAALARAAEAIGELPEYQRCLDLLDDCDPTARAELGLN
MSMEG_0754|M.smegmatis_MC2_155 FSHEPNQGFLRCVAALARAADDIGETDEYQRCLDLLDDCDPTARRELGLG
TH_1642|M.thermoresistible__bu YSHEPNRGFLRCVAALARAAEAIGETSEYQRCLDLLDDCDPGARAALGLA
MAB_4252c|M.abscessus_ATCC_199 YSHEPNRGFLRAVAALARAAKLIGEDHEYARCCDLLDDCDPAARPALLPA
::*:.*:****.******** *** ** ** :******* ** *
Rv0360c|M.tuberculosis_H37Rv --------
Mb0367c|M.bovis_AF2122/97 --------
MLBr_00284|M.leprae_Br4923 --------
MMAR_0672|M.marinum_M --------
MUL_2841|M.ulcerans_Agy99 --------
MAV_4783|M.avium_104 --------
Mflv_0232|M.gilvum_PYR-GCK --------
Mvan_0672|M.vanbaalenii_PYR-1 --------
MSMEG_0754|M.smegmatis_MC2_155 --------
TH_1642|M.thermoresistible__bu GDRESSGD
MAB_4252c|M.abscessus_ATCC_199 --------