For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARAVGIDLGTTNSVVSVLEGGDPVVVANSEGSRTTPSIVAFARNGEVLVGQPAKNQAVTNVDRTVRSVK RHMGSDWSIEIDGKKYTAPEISARILMKLKRDAEAYLGEDITDAVITTPAYFNDAQRQATKDAGQIAGLN VLRIVNEPTAAALAYGLDKGEKEQRILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLGGDDWDQRVVD WLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQSTSINLPYITVDADKNPLFLDEQLTRAEFQR ITQDLLDRTRKPFQSVIADTGISVSEIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTRLIERNTTIPTKRSETFTTADDNQPSVQIQVYQGERE IAAHNKLLGSFELTGIPPAPRGIPQIEVTFDIDANGIVHVTAKDKGTGKENTIRIQEGSGLSKEDIDRMI KDAEAHAEEDRKRREEADVRNQAETLVYQTEKFVKEQREAEGGSKVPEDTLNKVDAAVAEAKAALGGSDI SAIKSAMEKLGQESQALGQAIYEAAQAASQATGAAHPGGEPGGAHPGSADDVVDAEVVDDGREAK
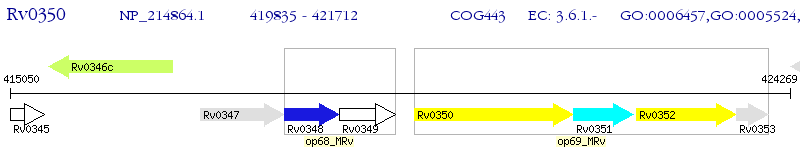
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0350 | dnaK | - | 100% (625) | molecular chaperone DnaK |
| M. tuberculosis H37Rv | Rv2264c | - | 3e-16 | 25.13% (382) | hypothetical protein Rv2264c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0358 | dnaK | 0.0 | 100.00% (625) | molecular chaperone DnaK |
| M. gilvum PYR-GCK | Mflv_0260 | dnaK | 0.0 | 90.72% (625) | molecular chaperone DnaK |
| M. leprae Br4923 | MLBr_02496 | dnaK | 0.0 | 92.48% (625) | molecular chaperone DnaK |
| M. abscessus ATCC 19977 | MAB_4273c | dnaK | 0.0 | 88.85% (628) | molecular chaperone DnaK |
| M. marinum M | MMAR_0637 | dnaK | 0.0 | 94.08% (625) | chaperone protein DnaK |
| M. avium 104 | MAV_4808 | dnaK | 0.0 | 92.80% (625) | molecular chaperone DnaK |
| M. smegmatis MC2 155 | MSMEG_0709 | dnaK | 0.0 | 89.52% (630) | molecular chaperone DnaK |
| M. thermoresistible (build 8) | TH_1023 | dnaK | 0.0 | 89.94% (626) | PROBABLE CHAPERONE PROTEIN DNAK (HEAT SHOCK PROTEIN 70) |
| M. ulcerans Agy99 | MUL_0593 | dnaK | 0.0 | 93.60% (625) | molecular chaperone DnaK |
| M. vanbaalenii PYR-1 | Mvan_0634 | dnaK | 0.0 | 90.08% (625) | molecular chaperone DnaK |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0260|M.gilvum_PYR-GCK MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
Mvan_0634|M.vanbaalenii_PYR-1 MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
Rv0350|M.tuberculosis_H37Rv MARAVGIDLGTTNSVVSVLEGGDPVVVANSEGSRTTPSIVAFARNGEVLV
Mb0358|M.bovis_AF2122/97 MARAVGIDLGTTNSVVSVLEGGDPVVVANSEGSRTTPSIVAFARNGEVLV
MMAR_0637|M.marinum_M MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
MUL_0593|M.ulcerans_Agy99 MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
MLBr_02496|M.leprae_Br4923 MARAVGIDLGTTNSVVSVLEGGDPVVVANSEGSRTTPSTVAFARNGEVLV
MAV_4808|M.avium_104 MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSIVAFARNGEVLV
TH_1023|M.thermoresistible__bu MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGARTTPSVVAFARNGEVLV
MSMEG_0709|M.smegmatis_MC2_155 MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
MAB_4273c|M.abscessus_ATCC_199 MARAVGIDLGTTNSVVAVLEGGDPVVVANSEGSRTTPSVVAFARNGEVLV
****************:***************:***** ***********
Mflv_0260|M.gilvum_PYR-GCK GQPAKNQAVTNVDRTIRSVKREIGTDWSVEIDGKNYTPQEISARVLMKLK
Mvan_0634|M.vanbaalenii_PYR-1 GQPAKNQAVTNVDRTIRSVKRHMGSDWTVKIDDKDYTPQEISARVLMKLK
Rv0350|M.tuberculosis_H37Rv GQPAKNQAVTNVDRTVRSVKRHMGSDWSIEIDGKKYTAPEISARILMKLK
Mb0358|M.bovis_AF2122/97 GQPAKNQAVTNVDRTVRSVKRHMGSDWSIEIDGKKYTAPEISARILMKLK
MMAR_0637|M.marinum_M GQPAKNQAVTNVDRTIRSVKRHMGGDWSIEIDDKKYTAPEISARVLMKLK
MUL_0593|M.ulcerans_Agy99 GQPAKNQAVTNVERTMRSVKRHMGGDWSIEIDDKKYTTPEISARVLMKLK
MLBr_02496|M.leprae_Br4923 GQPAKNQAVTNVDRTIRSVKRHMGSDWSIEIDGKKYTAQEISARVLMKLK
MAV_4808|M.avium_104 GQPAKNQAVTNVDRTIRSVKRHMGTDWSIEIDGKKYTAQEISARVLMKLK
TH_1023|M.thermoresistible__bu GQPAKNQAVTNVDRTVRSVKRHMGSDWTIDIDGKKYTPQEISARVLMKLK
MSMEG_0709|M.smegmatis_MC2_155 GQPAKNQAVTNVDRTIRSVKRHVGTDWNIEIDDKKYTPQEISARVLMKLK
MAB_4273c|M.abscessus_ATCC_199 GQPAKNQAVTNVDRTIRSVKRHMGTDWTVEIDGKNYTPQEISARTLQKLK
************:**:*****.:* **.:.**.*.**. ***** * ***
Mflv_0260|M.gilvum_PYR-GCK RDAESYLGEDITDAVITVPAYFNDAQRQATKDAGQIAGLNVLRIVNEPTA
Mvan_0634|M.vanbaalenii_PYR-1 RDAESYLGEDITDAVITVPAYFNDAQRQATKEAGQIAGLNVLRIVNEPTA
Rv0350|M.tuberculosis_H37Rv RDAEAYLGEDITDAVITTPAYFNDAQRQATKDAGQIAGLNVLRIVNEPTA
Mb0358|M.bovis_AF2122/97 RDAEAYLGEDITDAVITTPAYFNDAQRQATKDAGQIAGLNVLRIVNEPTA
MMAR_0637|M.marinum_M RDAEAYLGEDIADAVITVPAYFNDAQRQATKDAGQIAGLNVLRIVNEPTA
MUL_0593|M.ulcerans_Agy99 RDAEAYLGEDIADAVITVPAYFNDAQRQATKDAGQIAGLNVLRIVNEPTA
MLBr_02496|M.leprae_Br4923 RDAEAYLGEDITDAVITTPAYFNDAQRQATKEAGQIAGLNVLRIVNEPTA
MAV_4808|M.avium_104 RDAEAYLGEDITDAVITVPAYFNDAQRQATKEAGQIAGLNVLRIVNEPTA
TH_1023|M.thermoresistible__bu RDAEAYLGEDITDAVITVPAYFNDAQRQATKEAGQIAGMNVLRIVNEPTA
MSMEG_0709|M.smegmatis_MC2_155 RDAESYLGEDITDAVITVPAYFNDAQRQATKEAGQIAGLNVLRIVNEPTA
MAB_4273c|M.abscessus_ATCC_199 RDAEAYLGEDITDAVITVPAYFNDAQRQATKEAGQIAGLNVLRIVNEPTA
****:******:*****.*************:******:***********
Mflv_0260|M.gilvum_PYR-GCK AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
Mvan_0634|M.vanbaalenii_PYR-1 AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
Rv0350|M.tuberculosis_H37Rv AALAYGLDKGEKEQRILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
Mb0358|M.bovis_AF2122/97 AALAYGLDKGEKEQRILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
MMAR_0637|M.marinum_M AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
MUL_0593|M.ulcerans_Agy99 AALAYGLDKGEKDQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
MLBr_02496|M.leprae_Br4923 AALAYGLDKGEREQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
MAV_4808|M.avium_104 AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGEGVVEVRATSGDNHLG
TH_1023|M.thermoresistible__bu AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGDGVVEVRATSGDNHLG
MSMEG_0709|M.smegmatis_MC2_155 AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGDGVVEVRATSGDNHLG
MAB_4273c|M.abscessus_ATCC_199 AALAYGLDKGEKEQTILVFDLGGGTFDVSLLEIGDGVVEVRATSGDNHLG
***********::* *******************:***************
Mflv_0260|M.gilvum_PYR-GCK GDDWDDRIVEWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
Mvan_0634|M.vanbaalenii_PYR-1 GDDWDDRIVEWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
Rv0350|M.tuberculosis_H37Rv GDDWDQRVVDWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
Mb0358|M.bovis_AF2122/97 GDDWDQRVVDWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
MMAR_0637|M.marinum_M GDDWDDRVVEWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
MUL_0593|M.ulcerans_Agy99 GDDWDDRVVEWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
MLBr_02496|M.leprae_Br4923 GDDWDDRIVNWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
MAV_4808|M.avium_104 GDDWDDRIVNWLVDKFKGTSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
TH_1023|M.thermoresistible__bu GDDWDDRIVEWLVDKFKQSTGIDLTKDKMAMQRLREAAEKAKIELSASQQ
MSMEG_0709|M.smegmatis_MC2_155 GDDWDDRIVTWLVDKFKGSSGIDLTKDKMAMQRLREAAEKAKIELSSSQS
MAB_4273c|M.abscessus_ATCC_199 GDDWDERIVTWLVDKFKASAGIDLTKDKMAMQRLREAAEKAKIELSSSQS
*****:*:* ******* ::**************************:**.
Mflv_0260|M.gilvum_PYR-GCK TSINLPYITVDADKNPLFLDEQLTRSEFQKITQDLLDRTRKPFQSVIKDA
Mvan_0634|M.vanbaalenii_PYR-1 TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQQVIKDA
Rv0350|M.tuberculosis_H37Rv TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQSVIADT
Mb0358|M.bovis_AF2122/97 TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQSVIADT
MMAR_0637|M.marinum_M TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQSVIADT
MUL_0593|M.ulcerans_Agy99 TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQSVIADT
MLBr_02496|M.leprae_Br4923 TSVNLPYITVDSDKNPLFLDEQLIRAEFQRITQDLLDRTRQPFQSVVKDA
MAV_4808|M.avium_104 TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRQPFKSVIADA
TH_1023|M.thermoresistible__bu TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRKPFQQVIKDA
MSMEG_0709|M.smegmatis_MC2_155 TSINLPYITVDADKNPLFLDEQLTRAEFQRITQDLLDRTRQPFQQVIKDA
MAB_4273c|M.abscessus_ATCC_199 TSINLPYITVDADKNPLFLDEQLTRAEFQKITQDLLDRTRKPFQSVIKDA
**:********:*********** *:***:**********:**:.*: *:
Mflv_0260|M.gilvum_PYR-GCK GISVGEIDHVVLVGGSTRMPAVSELVKEMTGGKEPNKGVNPDEVVAVGAA
Mvan_0634|M.vanbaalenii_PYR-1 GIAVGDIDHVVLVGGSTRMPAVTELVKEMTGGQEPNKGVNPDEVVAVGAA
Rv0350|M.tuberculosis_H37Rv GISVSEIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
Mb0358|M.bovis_AF2122/97 GISVSEIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
MMAR_0637|M.marinum_M GISVSDIDHVVLVGGSTRMPAVTELVKELTGGKEPNKGVNPDEVVAVGAA
MUL_0593|M.ulcerans_Agy99 GISVSDIDHVVLVGGSTRMPAVTELVKELTGGKEPNKGVNPDEVVAVGAA
MLBr_02496|M.leprae_Br4923 GISVSEIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
MAV_4808|M.avium_104 GISVSDIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
TH_1023|M.thermoresistible__bu GISVSDIDHVVLVGGSTRMPAVSDLVKELTGGKEPNKGVNPDEVVAVGAA
MSMEG_0709|M.smegmatis_MC2_155 GISVSDIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
MAB_4273c|M.abscessus_ATCC_199 GVSVSDIDHVVLVGGSTRMPAVTDLVKELTGGKEPNKGVNPDEVVAVGAA
*::*.:****************::****:***:*****************
Mflv_0260|M.gilvum_PYR-GCK LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
Mvan_0634|M.vanbaalenii_PYR-1 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
Rv0350|M.tuberculosis_H37Rv LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTRLIERNTTIPTKRSETF
Mb0358|M.bovis_AF2122/97 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTRLIERNTTIPTKRSETF
MMAR_0637|M.marinum_M LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
MUL_0593|M.ulcerans_Agy99 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
MLBr_02496|M.leprae_Br4923 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
MAV_4808|M.avium_104 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
TH_1023|M.thermoresistible__bu LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSEVF
MSMEG_0709|M.smegmatis_MC2_155 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
MAB_4273c|M.abscessus_ATCC_199 LQAGVLKGEVKDVLLLDVTPLSLGIETKGGVMTKLIERNTTIPTKRSETF
*********************************:**************.*
Mflv_0260|M.gilvum_PYR-GCK TTADDNQPSVQIQVFQGEREIASHNKLLGSFELTGIPPAPRGVPQIEVTF
Mvan_0634|M.vanbaalenii_PYR-1 TTADDNQPSVQIQVFQGEREIAAHNKLLGSFELTGIPPAPRGVPQIEVTF
Rv0350|M.tuberculosis_H37Rv TTADDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGIPQIEVTF
Mb0358|M.bovis_AF2122/97 TTADDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGIPQIEVTF
MMAR_0637|M.marinum_M TTADDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGVPQIEVTF
MUL_0593|M.ulcerans_Agy99 TTADDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGVSQIEVTF
MLBr_02496|M.leprae_Br4923 TTADDNQPSVQIQVYQGEREIASHNKLLGSFELTGIPPAPRGVPQIEVTF
MAV_4808|M.avium_104 TTADDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGVPQIEVTF
TH_1023|M.thermoresistible__bu TTAEDNQPSVQIQVYQGEREIAAHNKLLGSFELTGIPPAPRGVPQIEVTF
MSMEG_0709|M.smegmatis_MC2_155 TTADDNQPSVQIQVYQGEREIASHNKLLGSFELTGIPPAPRGVPQIEVTF
MAB_4273c|M.abscessus_ATCC_199 TTADDNQPSVQIQVYQGEREIASHNKLLGSFELTGIPPAPRGVPQIEVTF
***:**********:*******:*******************:.******
Mflv_0260|M.gilvum_PYR-GCK DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMIKDAEAHADED
Mvan_0634|M.vanbaalenii_PYR-1 DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMIKDAEAHADED
Rv0350|M.tuberculosis_H37Rv DIDANGIVHVTAKDKGTGKENTIRIQEGSGLSKEDIDRMIKDAEAHAEED
Mb0358|M.bovis_AF2122/97 DIDANGIVHVTAKDKGTGKENTIRIQEGSGLSKEDIDRMIKDAEAHAEED
MMAR_0637|M.marinum_M DIDANGIVHVTAKDKGTGKENTIRIQEGSGLSKDEIDRMIKDAEAHAAED
MUL_0593|M.ulcerans_Agy99 DIDANGIVHVTAKDKGTGKENTIRIQEGSGLSKDEIDRMIKDAEAHAAED
MLBr_02496|M.leprae_Br4923 DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMVKDAEAHAEED
MAV_4808|M.avium_104 DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMIKDAEAHAEED
TH_1023|M.thermoresistible__bu DIDANGIVHVTAKDKGTGKENTIRIQEGSGLSQEEIERMIKDAEAHAEED
MSMEG_0709|M.smegmatis_MC2_155 DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMIKDAEAHAEED
MAB_4273c|M.abscessus_ATCC_199 DIDANGIVHVTAKDKGTGKENTIKIQEGSGLSKEEIDRMIKDAEAHADED
***********************:********:::*:**:******* **
Mflv_0260|M.gilvum_PYR-GCK RKRREEADVRNQAESLVYQTEKFVKEQREAE---GGSKVP-EDTLTKVDA
Mvan_0634|M.vanbaalenii_PYR-1 RQRREEADVRNQAESLVYQTEKFVKEQREAE---GGSKVP-EDTLTKVDS
Rv0350|M.tuberculosis_H37Rv RKRREEADVRNQAETLVYQTEKFVKEQREAE---GGSKVP-EDTLNKVDA
Mb0358|M.bovis_AF2122/97 RKRREEADVRNQAETLVYQTEKFVKEQREAE---GGSKVP-EDTLNKVDA
MMAR_0637|M.marinum_M HKRREEADVRNQAETLVYQTEKFVKEQREAE---GGSKVP-EDTLNKVDA
MUL_0593|M.ulcerans_Agy99 HKRREEADVRNQAETLVYQTEKFVKEQREAE---GGSKVP-EDTLNKVDA
MLBr_02496|M.leprae_Br4923 RKRREEADVRNQAETLVYQTEKFVKEQRETE---NGSRVP-EDTLNKVEA
MAV_4808|M.avium_104 RKRREEADVRNQAESLVYQTEKFVKDQREAE---GGSKVP-EETLSKVDA
TH_1023|M.thermoresistible__bu RRRREEAEVRNQAETLVYQTEKFVKEQREAE---GGSKVP-EETLNKVEA
MSMEG_0709|M.smegmatis_MC2_155 RKRREEADVRNQAESLVYQTEKFVAEQRGAASDGGGSKVP-EETLAKVDS
MAB_4273c|M.abscessus_ATCC_199 KKRREEADVRNQAESLVYQTEKFVKEQREPAE--GSEKVVSDETLSKVDE
::*****:******:********* :** . ...:* ::** **:
Mflv_0260|M.gilvum_PYR-GCK AISDAKAALAGTDIAAIKSAMEKLGEESQALGQAIYEATQAEQAAGGAP-
Mvan_0634|M.vanbaalenii_PYR-1 AIADAKTALAGTDIGAIKSAMEKLGEESQALGQAIYEATQAEQAAGGAA-
Rv0350|M.tuberculosis_H37Rv AVAEAKAALGGSDISAIKSAMEKLGQESQALGQAIYEAAQAASQATGAAH
Mb0358|M.bovis_AF2122/97 AVAEAKAALGGSDISAIKSAMEKLGQESQALGQAIYEAAQAASQATGAAH
MMAR_0637|M.marinum_M AVAEAKSALAGTDIAAIKSAMEKLGEESQALGQAIYEATQADAAAAGA--
MUL_0593|M.ulcerans_Agy99 AVAEAKSALAGTDIAAIKSAMEKLGQESQALGQAIYEATQADAAAAGA--
MLBr_02496|M.leprae_Br4923 AVAEAKTALGGTDISAIKSAMEKLGQDSQALGQAIYEATQAASKVGG---
MAV_4808|M.avium_104 AIADAKTALGGTDITAIKSAMEKLGQESQALGQAIYEATQAESAQAGGPD
TH_1023|M.thermoresistible__bu AINDAKTALNGTDIGAIKSAMEKLGQESQALGQAIYEATQAAQAAGD---
MSMEG_0709|M.smegmatis_MC2_155 AIADAKKALEGTDISAIKSAMEKLGVESQALGQAIYEATQAEQPAGGS--
MAB_4273c|M.abscessus_ATCC_199 AISEAKKALEGTDIGAIKAAMEKLGEQSQALGQAIYEAAAAKAQADG---
*: :** ** *:** ***:****** :***********: * .
Mflv_0260|M.gilvum_PYR-GCK GGESTGASNSG--DDVVDAEVVEDDQ--ERK
Mvan_0634|M.vanbaalenii_PYR-1 GGDSAGASGAN--DDVVDAEVVEDDQ--ERK
Rv0350|M.tuberculosis_H37Rv PGGEPGGAHPGSADDVVDAEVVDDGR--EAK
Mb0358|M.bovis_AF2122/97 PGGEPGGAHPGSADDVVDAEVVDDGR--EAK
MMAR_0637|M.marinum_M PGGQPGGAEPG-ADDVVDAEVVDDDQ--ESK
MUL_0593|M.ulcerans_Agy99 PGGQPGGAEPG-ADDVVDAEVVDDDQ--ESK
MLBr_02496|M.leprae_Br4923 EASAPGGSNST--DDVVDAEVVDDER--ESK
MAV_4808|M.avium_104 GAAAGGGSGSA--DDVVDAEVVDDDR--ESK
TH_1023|M.thermoresistible__bu --GEAGPAGGG-ADDVVDAEVVDEER--ENK
MSMEG_0709|M.smegmatis_MC2_155 DNGAPGD------DNVVDAEVVDDDAGKENK
MAB_4273c|M.abscessus_ATCC_199 --AEGGAGSAD--SDVVDAEVVDEEK--DSK
* .:*******:: : *