For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TISVFDLFTIGIGPSSSHTVGPMRAANQFVVALRRRGHLDDLEAMRVDLFGSLAATGAGHGTMSAILLGL EGCQPETITTEHKERRLAEIAASGVTRIGGVIPVPLTERDIDLHPDIVLPTHPNGMTFTAAGPHGRVLAT ETYFSVGGGFIVTEQTSGNSGQHPCSVALPYVSAQELLDICDRLDVSISEAALRNETCCRTENEVRAALL HLRDVMVECEQRSIAREGLLPGGLRVRRRAKVWYDRLNAEDPTRKPEFAEDWVNLVALAVNEENASGGRV VTAPTNGAAGIVPAVLHYAIHYTSAGAGDPDDVTVRFLLTAGAIGSLFKERASISGAEVGCQGEVGSAAA MAAAGLAEILGGTPRQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERNAISAGKAINAARMALRGDGIHR VTLDQVIDTMRATGADMHTKYKETSAGGLAINVAVNIVEC*
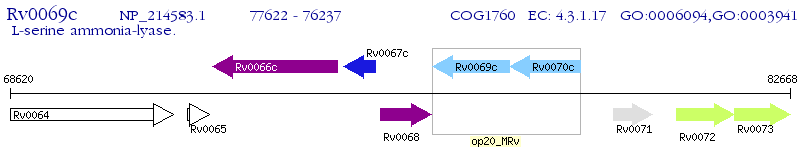
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0069c | sdaA | - | 100% (461) | PROBABLE L-SERINE DEHYDRATASE SDAA (L-SERINE DEAMINASE) (SDH) (L-SD) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0070c | sdaA | 0.0 | 100.00% (461) | L-serine dehydratase SdaA |
| M. gilvum PYR-GCK | Mflv_0819 | - | 1e-124 | 53.22% (466) | L-serine dehydratase 1 |
| M. leprae Br4923 | MLBr_01755 | sdaA | 1e-172 | 68.98% (461) | L-serine dehydratase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0207 | sdaA | 0.0 | 82.65% (461) | L-serine dehydratase SdaA |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3440 | sdaA | 0.0 | 70.74% (458) | L-serine ammonia-lyase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4901 | sdaA | 0.0 | 82.65% (461) | L-serine dehydratase SdaA |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0069c|M.tuberculosis_H37Rv MTISVFDLFTIGIGPSSSHTVGPMRAANQFVVALRRRGHLDDLEAMRVDL
Mb0070c|M.bovis_AF2122/97 MTISVFDLFTIGIGPSSSHTVGPMRAANQFVVALRRRGHLDDLEAMRVDL
MMAR_0207|M.marinum_M MTISVFELFTVGIGPSSSHTVGPMRAANEFIAALRSAGQRDAVAAVQIDL
MUL_4901|M.ulcerans_Agy99 MTISVFELFTVGIGPSSSHTVGPMRAANEFIAALRSAGQRDAVAAVQIDL
MSMEG_3440|M.smegmatis_MC2_155 ----MFDLFSVGIGPSSSHTVGPMRAAARFADELDAIGVLDAVATIDVDL
MLBr_01755|M.leprae_Br4923 MTISVFDLFTIGIGPSSSHTVGPMRAAARFVDGLAANGLLEDVCDIRVEL
Mflv_0819|M.gilvum_PYR-GCK MAVSVFDLFSIGIGPSSSHTVGPMRAARRFVGLLENDGSPAGVAGVQVEL
:*:**::**************** .* * * : : ::*
Rv0069c|M.tuberculosis_H37Rv FGSLAATGAGHGTMSAILLGLEGCQPETITTEHKERRLAEIAASGVTRIG
Mb0070c|M.bovis_AF2122/97 FGSLAATGAGHGTMSAILLGLEGCQPETITTEHKERRLAEIAASGVTRIG
MMAR_0207|M.marinum_M FGSLAATGAGHGTMSAVLLGLEGFQPEAISSEAKERRLAEIAESGVTRID
MUL_4901|M.ulcerans_Agy99 FGSLAATGAGHGTMSAVLLGLEGFQPEAISSEAKERRLAEIAESGVTRID
MSMEG_3440|M.smegmatis_MC2_155 YGSLAATGAGHGTMSAILLGLEGSRPETIESEFKDRRIEQMRAEGRIRVG
MLBr_01755|M.leprae_Br4923 FGSLAATGAGHGTMSAILLGLEGEHPERLDADDMVTRLACMRAAGTICLG
Mflv_0819|M.gilvum_PYR-GCK YGSLGATGAGHGTPGAVMLGLEGADPESVDPDEARHRVAEIGERRQLLLA
:***.******** .*::***** ** : .: *: : :
Rv0069c|M.tuberculosis_H37Rv GVIPVPLTERDIDLHPDIVLPTHPNGMTFTAAGPHGRVLATETYFSVGGG
Mb0070c|M.bovis_AF2122/97 GVIPVPLTERDIDLHPDIVLPTHPNGMTFTAAGPHGRVLATETYFSVGGG
MMAR_0207|M.marinum_M GTVAVPLTEGDVKLHPDVVLPTHPNGMTLTATDSGGRVLATETYFSVGGG
MUL_4901|M.ulcerans_Agy99 GTVAVPLTEGDVKLHPDVVLPTHPNGMTLTATDSGGRVLVTETYFSVGGG
MSMEG_3440|M.smegmatis_MC2_155 GRVEITLSENDIRLHPETVLPTHSNGMRLSAAGSDGRPLHAQTYFSVGGG
MLBr_01755|M.leprae_Br4923 GLIVIKFTEADVVLRPQTVLDRHPNAMTIVALARDGTKLAAETYYSVGGG
Mflv_0819|M.gilvum_PYR-GCK GEHAIAFDPAADIVLSLRAMDFHSNGMVFTAFDPIGAPVLRRVYYSVGGG
* : : : . .: *.*.* : * * : ..*:*****
Rv0069c|M.tuberculosis_H37Rv FIVTEQTSGNSGQHPCS-VALPYVSAQELLDICDRLDVSISEAALRNETC
Mb0070c|M.bovis_AF2122/97 FIVTEQTSGNSGQHPCS-VALPYVSAQELLDICDRLDVSISEAALRNETC
MMAR_0207|M.marinum_M FIVTAGSDHSDDRHSCR-TPLPYSSGQELLDICDQFGISISEVALRNESC
MUL_4901|M.ulcerans_Agy99 FIVTAGSDHSDDRHSCR-TPLPYSSGQELLDICDQFGISISEVALRNESC
MSMEG_3440|M.smegmatis_MC2_155 FIVAEGADELGQGHGSEDGDVTFSSAAELLALADKFDSPISSVMLAFETE
MLBr_01755|M.leprae_Br4923 FVVAHGDPPEGAEWTTG--GVSFGSAGELLELAATRGMSICELMAEYEQS
Mflv_0819|M.gilvum_PYR-GCK FVVDEDEAAEDAPDQVA-LPFPFHTGAELVRLAEENRCRIADLVSANEQA
*:* . ..: :. **: :. *.. *
Rv0069c|M.tuberculosis_H37Rv CRTENEVRAALLHLRDVMVECEQRSIAREGLLPGGLRVRRRAKVWYDRLN
Mb0070c|M.bovis_AF2122/97 CRTENEVRAALLHLRDVMVECEQRSIAREGLLPGGLRVRRRAKVWYDRLN
MMAR_0207|M.marinum_M CRSAQDVRSGLLHLRDVMVECEQQSISRDGLLPGSLQVRRRAKSWYERLN
MUL_4901|M.ulcerans_Agy99 CRSAQDVRSGLLHLRDVMVECEQQSISRDGLLPGSLQVRRRAKSWYERLN
MSMEG_3440|M.smegmatis_MC2_155 TRSEDEVRARLLHIRDVMFECMERGITRDGFLPGTLRVRRRARDWYQRLL
MLBr_01755|M.leprae_Br4923 LRSEQEVRAGLLNIRDVMTQSVQRGIAQGGYLPGGLKVRRRAHSWHERLC
Mflv_0819|M.gilvum_PYR-GCK LGNGARLRTGLLRIWSAMRDCVARGLATDGTLPGSLHVQRRAHVLAEALE
. :*: **.: ..* :. :.:: * *** *:*:***: : *
Rv0069c|M.tuberculosis_H37Rv AEDPTRKPEFAEDWVNLVALAVNEENASGGRVVTAPTNGAAGIVPAVLHY
Mb0070c|M.bovis_AF2122/97 AEDPTRKPEFAEDWVNLVALAVNEENASGGRVVTAPTNGAAGIVPAVLHY
MMAR_0207|M.marinum_M AEDPNRKPEFAEDWVNLVALAVNEENASGGRVVTAPTNGAAGIVPAVLHY
MUL_4901|M.ulcerans_Agy99 AEDPNRKPEFAEDWVNLVALAVNEENASGGRVVTAPTNGAAGIVPAVLHY
MSMEG_3440|M.smegmatis_MC2_155 TDDPGRDPVYAEDWVNLVALAVNEENASGGRIVTAPTNGAAGIIPAVLYY
MLBr_01755|M.leprae_Br4923 AEDPNRDPVFAEDWVNLVALAVNEENAVGGRVVTAPTNGAAGIIPAVLHY
Mflv_0819|M.gilvum_PYR-GCK RD--RSDPLYAMDWVTVYALAVNEENAAGGRVVTAPTNGAAGIIPAVLHY
: .* :* ***.: ********* ***:***********:****:*
Rv0069c|M.tuberculosis_H37Rv AIHYTSAGAGDPDDVTVRFLLTAGAIGSLFKERASISGAEVGCQGEVGSA
Mb0070c|M.bovis_AF2122/97 AIHYTSAGAGDPDDVTVRFLLTAGAIGSLFKERASISGAEVGCQGEVGSA
MMAR_0207|M.marinum_M AAHYTRAGTADSEDATVRFLLTAGAIGSLFKERASISGAEVGCQGEVGSA
MUL_4901|M.ulcerans_Agy99 AAHYTRAGTADSDDATVRFLLTAGAIGSLFKERASISGAEVGCQGEVGSA
MSMEG_3440|M.smegmatis_MC2_155 ALHYTPQGAADPDDASVRFLLTAGAIGSLYKERASISGAEVGCQGEVGSA
MLBr_01755|M.leprae_Br4923 AQHYCPAGRADPDDTSVRFLLTAGAIGSLYKELASISGAEVGCQGEVGSA
Mflv_0819|M.gilvum_PYR-GCK YWRFVPEAD---EDGVVEFLLTAAAIGQLFKANASISGAEVGCQGEVGSA
:: . :* *.*****.***.*:* *****************
Rv0069c|M.tuberculosis_H37Rv AAMAAAGLAEILGGTPRQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERN
Mb0070c|M.bovis_AF2122/97 AAMAAAGLAEILGGTPRQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERN
MMAR_0207|M.marinum_M AAMAAAGLAEVLGGTPRQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERN
MUL_4901|M.ulcerans_Agy99 AAMAAAGLAEVLGGTPRQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERN
MSMEG_3440|M.smegmatis_MC2_155 ASMAAAGLAEILGGTPAQVENAAEIAMEHSLGLTCDPIGGLVQIPCIERN
MLBr_01755|M.leprae_Br4923 ASMAAAGLAEILGGTPAQVENAAEIAMEHSLGLTCDPIAGLVQIPCIERN
Mflv_0819|M.gilvum_PYR-GCK CSMAAGALAAVLGGTPAQVENAAEIGIEHNLGLTCDPVGGLVQIPCIERN
.:***..** :***** ********.:**.*******:.***********
Rv0069c|M.tuberculosis_H37Rv AISAGKAINAARMALRGDGIHRVTLDQVIDTMRATGADMHTKYKETSAGG
Mb0070c|M.bovis_AF2122/97 AISAGKAINAARMALRGDGIHRVTLDQVIDTMRATGADMHTKYKETSAGG
MMAR_0207|M.marinum_M AISAGKAINAARMALRGDGFHRVTLDQVIDTMRATGADMHAKYKETSAGG
MUL_4901|M.ulcerans_Agy99 AISAGKAINAARMALRGDGFHRVTLDQVIDTMRATGADMHAKYKETSAGG
MSMEG_3440|M.smegmatis_MC2_155 AISAGKAINAARMALRGDGTHRVSLDQVIETMRSTGRDMSAKYKETSTGG
MLBr_01755|M.leprae_Br4923 AISAGKAINAARMAMRGDGTHRVSLDEVIATMRATGRDMNFKYKETAAGG
Mflv_0819|M.gilvum_PYR-GCK AIASVKAIAAARLALFGDGTHRVSLDTAIKTMRDTGADMAEKYKETSLGG
**:: *** ***:*: *** ***:** .* *** ** ** *****: **
Rv0069c|M.tuberculosis_H37Rv LAINVAVNIVEC
Mb0070c|M.bovis_AF2122/97 LAINVAVNIVEC
MMAR_0207|M.marinum_M LAINVAVNIVEC
MUL_4901|M.ulcerans_Agy99 LAINVAVNIVEC
MSMEG_3440|M.smegmatis_MC2_155 LATAVPVNVVEC
MLBr_01755|M.leprae_Br4923 LAL--AVNIVEC
Mflv_0819|M.gilvum_PYR-GCK LAL----NVVEC
** *:***