For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTATVRWLTVLLLVGAVLAGVGRSASEHRPDDKLIEGPYASLLDASTDLGPAREQSIEFTASLTDRSRP TALIDWARDRSLSVRWRPGDDWVVVEGSPDAVTQAFDVSVRDYRGRMGQLFYASPHQPAVPEHLRAEVSE MGRILSFIPHRMSRPDHIPLNVPDRGLGPDALLNAYNADDLARAGFTGKGITIVVFAFDGFRQSDLDTFT TMFGLPQFTPEIVGGSPGEPRGELSMDLQVAHAIAPDARKVVVNARPTVEDGGGYRKIGEMLEDTDRRFP GAVWSFSIGWGCDKLITAADLAPVRSALRTAQSHGTTAYNASGDLAGMECRGGQDWSSPPSEQDIGLDSI ASLPEMTSVGGTTLSTDADGTWVSEQTWYDVPLSQGTGGGVSALFDMPAWQRTAAGAVPPERNTGKRMTP DVAAVADPFTGVKIVLDDQVLVGGGTSQSAPIWAGLTAVMNQYLLEHNGTLIGDINPLLYRIAEGAPRPA YRDVTLGGNAVDNAGPGYDLVTGLGTPDIDNLVRNLQLAQKVQA
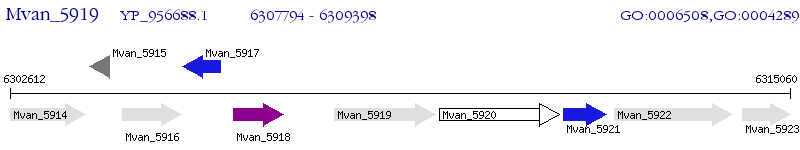
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5919 | - | - | 100% (534) | kumamolisin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1617 | - | 0.0 | 66.35% (529) | kumamolisin |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5919|M.vanbaalenii_PYR-1 MTTAT-----------------VRWLTVLLLVGAVLAGVGRSAS-EHRPD
MSMEG_1617|M.smegmatis_MC2_155 MAGTTRHVGVSIRNPGGFRHWHTHWSLALIMIGALLISVFRITSGPDGHG
*: :* .:* .*:::**:* .* * :* . .
Mvan_5919|M.vanbaalenii_PYR-1 DKLIEGPYASLLDASTDLGPAREQSIEFTASLTDRSRPTALIDWARDRSL
MSMEG_1617|M.smegmatis_MC2_155 PAEIAGPYAHLLAASTDLGPADGGPVQLTVTLD--GSPEALTSWARERGL
* **** ** ******** .:::*.:* . * ** .***:*.*
Mvan_5919|M.vanbaalenii_PYR-1 SVRWRPGDDWVVVEGSPDAVTQAFDVSVRDYRGRMGQLFYASPHQPAVPE
MSMEG_1617|M.smegmatis_MC2_155 SVRWRHGDTWAIVEGPPADMAAAFEVDIHDYRGRRGQEFYASPQQPSIPE
***** ** *.:***.* :: **:*.::***** ** *****:**::**
Mvan_5919|M.vanbaalenii_PYR-1 HLRAEVSEMGRILSFIPHRMSRPD--HIPLNVPDRGLGPDALLNAYNADD
MSMEG_1617|M.smegmatis_MC2_155 PVRSEVTEVGRILGYTPYRMNTPDLQHLPAEVPDRGLTPQLLLNTYNLRG
:*:**:*:****.: *:**. ** *:* :****** *: ***:** .
Mvan_5919|M.vanbaalenii_PYR-1 LARAGFTGKGITIVVFAFDGFRQSDLDTFTTMFGLPQFTPEIVGGSPGEP
MSMEG_1617|M.smegmatis_MC2_155 LAEQGYTGKGQTIVIFAFDGFDQADLDTYADTFDLPRFTPILVGGQPSTP
**. *:**** ***:****** *:****:: *.**:*** :***.*. *
Mvan_5919|M.vanbaalenii_PYR-1 RGELSMDLQVAHAIAPDARKVVVNARPTVEDGGGYRKIGEMLEDTDRRFP
MSMEG_1617|M.smegmatis_MC2_155 RGETTMDLQVAHAIAPDARKVVVNARPTVQGDGAYEKIGQMLEDADRQFP
*** :************************:..*.*.***:****:**:**
Mvan_5919|M.vanbaalenii_PYR-1 GAVWSFSIGWGCDKLITAADLAPVRSALRTAQSHGTTAYNASGDLAGMEC
MSMEG_1617|M.smegmatis_MC2_155 GAVWSFSIGWGCDKLITAADLAPVRSALAAAHTRGTTAFNASGDLAGLEC
**************************** :*:::****:********:**
Mvan_5919|M.vanbaalenii_PYR-1 RGGQDWSSPPSEQDIGLDSIASLPEMTSVGGTTLSTDADGTWVSEQTWYD
MSMEG_1617|M.smegmatis_MC2_155 KGGQDWSSAPGDDDIGLDSVASLPEITDVGGTTLSTTADGAWLAEQAWFD
:*******.*.::******:*****:*.******** ***:*::**:*:*
Mvan_5919|M.vanbaalenii_PYR-1 VPLSQGTGGGVSALFDMPAWQRTAAGAVPPERNTGKRMTPDVAAVADPFT
MSMEG_1617|M.smegmatis_MC2_155 VPLSQGTGGGVSALFDRPDWQQGFS--APDDGGAQRRLTPDIAAVADPFT
**************** * **: : .* : .: :*:***:********
Mvan_5919|M.vanbaalenii_PYR-1 GVKIVLDDQVLVGGGTSQSAPIWAGLTAVMNQYLLEHNGTLIGDINPLLY
MSMEG_1617|M.smegmatis_MC2_155 GVKIVLNNQVLVGGGTSQAAPLWAGIAAVMNQYLVANGGRAIGDLNPMLY
******::**********:**:***::*******: :.* ***:**:**
Mvan_5919|M.vanbaalenii_PYR-1 RIAEGAPRPAYRDVTLGGNAVDNAGPGYDLVTGLGTPDIDNLVRNLQLAQ
MSMEG_1617|M.smegmatis_MC2_155 QIARGAPLPAFRDVTLGGNAVATAGPGYDLVTGLGTPDTENLVKNLLVLQ
:**.*** **:********** .*************** :***:** : *
Mvan_5919|M.vanbaalenii_PYR-1 KVQA
MSMEG_1617|M.smegmatis_MC2_155 KANP
*.:.