For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNADLIDTVATAGAAIEQAVAVFMLHPETYGGSIAAGYQNPLAGYVAGRGGVLGEATGATVSAVFAVFEP VGLAAMWDEGVAVHGAARASEIYWDQTAEFGRKYLSGAEGLDRIAALGEKLIASTPITALPLFAGWRAMP FAEDAPARALQVMFVLRELRAGLHFNALSLSGLTPVEAHMLNKGPGYTAMFGWPEPFADGADKKDGYAEI EQATNRRIADIFSAALDPAEAEELARLSTAALASLKANVPA*
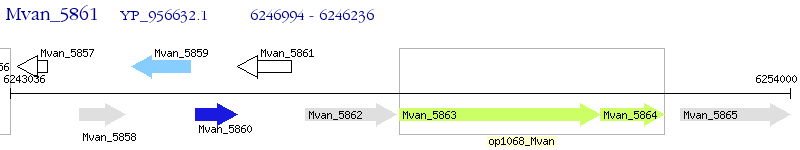
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5861 | - | - | 100% (252) | hypothetical protein Mvan_5861 |
| M. vanbaalenii PYR-1 | Mvan_3405 | - | 2e-09 | 29.14% (175) | hypothetical protein Mvan_3405 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0977 | - | 1e-119 | 82.54% (252) | hypothetical protein Mflv_0977 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2220c | - | 6e-05 | 24.84% (161) | hypothetical protein MAB_2220c |
| M. marinum M | MMAR_0126 | - | 1e-101 | 71.08% (249) | hypothetical protein MMAR_0126 |
| M. avium 104 | MAV_0134 | - | 4e-11 | 28.65% (171) | hypothetical protein MAV_0134 |
| M. smegmatis MC2 155 | MSMEG_6750 | - | 1e-109 | 76.02% (246) | hypothetical protein MSMEG_6750 |
| M. thermoresistible (build 8) | TH_0197 | - | 1e-106 | 72.22% (252) | hypothetical protein MSMEG_6750 |
| M. ulcerans Agy99 | MUL_3188 | - | 3e-06 | 25.75% (167) | hypothetical protein MUL_3188 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5861|M.vanbaalenii_PYR-1 ------------------MSNADLIDTVATAGAAIEQAVAVFMLHPETYG
Mflv_0977|M.gilvum_PYR-GCK --MSPAHNERGPGATLPSMSNADLIESVAAAGTAIEQAVAVFMLHPETYG
TH_0197|M.thermoresistible__bu ------------------MSDTDMTKAVADAGAAMEQAVALFMLHPETFS
MSMEG_6750|M.smegmatis_MC2_155 MRHAPTITDDCRHQEQRYMS-TTVLDATDAAGSAIEEAVAVFMLHPETFA
MMAR_0126|M.marinum_M ------------------MSDSDMFDAARAAGSPIEQAVAVFMLHPDTFA
MAV_0134|M.avium_104 --------------------MRRQPLLARRFFDRYEPVHAVTYFAPEARA
MUL_3188|M.ulcerans_Agy99 --------------------MQRTPELARRLYDRFEPIHAVTYFAPEARA
MAB_2220c|M.abscessus_ATCC_199 --------------------MARSAVLARRLYERFEPVHGIVYFAPEARA
. * .: : *:: .
Mvan_5861|M.vanbaalenii_PYR-1 GSIAAGYQNPLAGYVAGRGGVLGEATGATVSAVFAVFEPVGLAAMWDEGV
Mflv_0977|M.gilvum_PYR-GCK GSVAAGYENPLAGYIAGRGGVLGEANGATVAAVFAVFEPTAVAAMWDEGV
TH_0197|M.thermoresistible__bu ASQAAGYQNPLAGYVAGRGGVLGQASGATVASVFVVFEPTGLAALWDEGV
MSMEG_6750|M.smegmatis_MC2_155 ESVAAGYQNPLAGYVAGRGGVLGDTTGATVAAVFAVFEPAGLSALWDEGV
MMAR_0126|M.marinum_M ESIEAGYEHPFAGYVAGRAGVLGETTGVTVHSVFAVFEPSFIHGMWETGI
MAV_0134|M.avium_104 ALDALGYRGFWMGYFAARSAPLGAVPREVVAAIFYNFAPERVAKALPAAW
MUL_3188|M.ulcerans_Agy99 ALANLGYRGFWMGYFAARSAPLGQVAPEVVTAAFYNFAPARVAKALPAAW
MAB_2220c|M.abscessus_ATCC_199 AVEDLGYRGFWRGYFATRSAPLGRADARVVQAIFYNFASFRVRKSLDGAW
**. **.* *.. ** . .* : * * . : .
Mvan_5861|M.vanbaalenii_PYR-1 AVHGAARASEIYWDQTAEFGRKY-LSGAEGLDRIAALGEKLIASTPITAL
Mflv_0977|M.gilvum_PYR-GCK AVRGAAGAAEVYWNQTAEFGRKY-LAGADGLDRIAALGEKLIASAPNWGV
TH_0197|M.thermoresistible__bu AVHGAEGAAQRYWAQTADFGRKY-LSGADGLDRLAGLGEKVIAAAPPAGV
MSMEG_6750|M.smegmatis_MC2_155 AVRGAAGAAQVYWEQAAAFGRKY-LAGAEGLDRLAALGEKVVAVTPTVAL
MMAR_0126|M.marinum_M AVRGAIGAAELYWDQVAGFARKY-LAAATGLDRIAALGEKIIAATPDAGL
MAV_0134|M.avium_104 EIAGPQAALRARQESAVAALRRYGLRPDENVAVAAELAAKAARGAPLDGR
MUL_3188|M.ulcerans_Agy99 DIAEPRLALQARQDSAVAALRRYGLTDDDNVRSAGRLAGSAASRASLEGR
MAB_2220c|M.abscessus_ATCC_199 DSISPAQALAVRQAGAVAALRRYGLTDDENLRTAAELLGRAAHEASSSGR
. * .. *:* * .: . * :. .
Mvan_5861|M.vanbaalenii_PYR-1 PLFAGWRAMPFAEDAPARALQVMFVLRELRAGLHFNALSLSGLTPVEAHM
Mflv_0977|M.gilvum_PYR-GCK PLFVGWRAMPLAEDSPARALQVMFVLRELRAALHFNALSLSGVTPIEAHM
TH_0197|M.thermoresistible__bu PLFAGWRTMPLADDAPARALQVMFVLRELRAGLHFNALTLSDISPIEAHM
MSMEG_6750|M.smegmatis_MC2_155 PLFAGWRTMPLAGDAPARALQVMFVLRELRGGVHFNLLALSGVTPVEAHM
MMAR_0126|M.marinum_M PLYAGWRTMALADDAAARALQVMFVLRELRAGVHFNALSISGITPVEAHM
MAV_0134|M.avium_104 PLFAANLALPWPEDPLAALWHATTLLREQRGDAHVAVLAAAGISGRESNV
MUL_3188|M.ulcerans_Agy99 ALFGANRALPWPEDPVAALWHATTLLREHRGDGHVAVLVAAGVSGREPNV
MAB_2220c|M.abscessus_ATCC_199 PLYAALTTVPWPDEPVAALWHAATLLREQRGDGHVAALVAAGIGGRESNV
.*: . ::. . :. * :. :*** *. *. * :.: *.::
Mvan_5861|M.vanbaalenii_PYR-1 LNKGPG--YTAMFGWPEPFADG----------------------ADKKDG
Mflv_0977|M.gilvum_PYR-GCK LNKGPQ--YAAMFGWPEPYADG----------------------ADKKDR
TH_0197|M.thermoresistible__bu LNKGRQ--YAQMFGWPEPYPEG----------------------TDKKDR
MSMEG_6750|M.smegmatis_MC2_155 LNKGPQ--YTAVFGWPEPYADG----------------------ADKLDR
MMAR_0126|M.marinum_M LNKGHE--YTTMFGWPEPFADG----------------------TDKKDR
MAV_0134|M.avium_104 LHAAAGNVSRDYIARTRDYDEASWRRHEQRLAERGLLDGDGALTAAGRQC
MUL_3188|M.ulcerans_Agy99 FHCAADGVPNDYMKLTRHYDDAEWQACVNSLVGRGLLTEEGSLAPAGREL
MAB_2220c|M.abscessus_ATCC_199 FHVASGRATKDTIMRSRDYDEEEWGEVEAELTRRGLLEG-GGLSPYGAAV
:: . : .. : : .
Mvan_5861|M.vanbaalenii_PYR-1 YAEIEQATNRRIADIFSAALDPAEAEELARLSTAALASLKANVPA-----
Mflv_0977|M.gilvum_PYR-GCK YDEIEQATNRRMAELFSAALENGEAGELARLSTAALATLKANVPS-----
TH_0197|M.thermoresistible__bu YAEVEQATNRRMAQIYASALTVAEAQELAQLSADALAAVKANAPS-----
MSMEG_6750|M.smegmatis_MC2_155 YDEIRQETNRRMSELLSAALDPAEVDELAALSAAALATLKANVPG-----
MMAR_0126|M.marinum_M YAAVEEATNRRMAEIFGAALDGGEADELARLSTAALATLKASAAS-----
MAV_0134|M.avium_104 KDHIEATTDALALSALDALTDDEVETLFEALTPITRAVIAGGDIPAATPM
MUL_3188|M.ulcerans_Agy99 HEHLENTTDALALRAFDGLDDAELDALFQALTQVAKTVISAGDLPSVTPM
MAB_2220c|M.abscessus_ATCC_199 KADIEDRTDRAFLPVLDVLTDAELETLFRSLTPVTRQVIAAGDLPASTPM
:. *: : *: : : ..
Mvan_5861|M.vanbaalenii_PYR-1 -----------------
Mflv_0977|M.gilvum_PYR-GCK -----------------
TH_0197|M.thermoresistible__bu -----------------
MSMEG_6750|M.smegmatis_MC2_155 -----------------
MMAR_0126|M.marinum_M -----------------
MAV_0134|M.avium_104 TLRRDELHDDSAHLVAR
MUL_3188|M.ulcerans_Agy99 SIRRDF-----------
MAB_2220c|M.abscessus_ATCC_199 GLRRNDLDDDSAHLT--