For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLLVPGDTCWRIEQADHFAGIVDGADYFRHVKAAMMRARHRVMLIGWDFDTRMTFERGDKTLAGPNQLRA FLWWLLRLRPDLEIYLLKSNLGLIPAFQTIWDGVTPVKLLNWLSSDRLWFAVDGAHPTGAVHHQKIVVVD DVLAFCGGIDLTVDRWDTSEHLQHNTIRRTMGRAYGPRHEVAVAVDGAAARALGDQARNRWRAATKQLLG PIEDADRIWPPGLEPSLRRVGVGIARTLPAQTPGGEVREVEALNRAAIAAGRRTIYLENQYLASRPLVSA LASRLQEPDGPEIVIVLPRRSESRLERESMDTARHDLLEILWAADDHHRLGVYWPVADGGAPVYVHSKVL VSDDRLLRIGSSNLNNRSMGFDSECDVAIEADPDDRDHDAVRRVITATRDGLVGEHLGVPAAEVASAVRE HGSFLNALRALRGPGRTLVEFTADTVRDEAGRLAENDLMDPDRVPRSITRTVQRLFGRLTE*
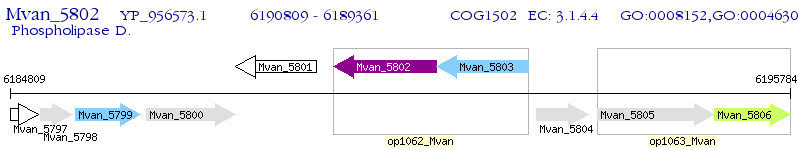
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5802 | - | - | 100% (482) | phospholipase D/transphosphatidylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1004 | - | 1e-171 | 64.26% (470) | phospholipase D/transphosphatidylase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3989 | - | 0.0 | 66.88% (474) | phospholipase D family protein |
| M. smegmatis MC2 155 | MSMEG_0375 | - | 9e-22 | 31.65% (357) | phospholipase D family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5802|M.vanbaalenii_PYR-1 -------MTLLVPGDTCWRIEQADHFAG-----IVDGADYFRHVKAAMMR
MAV_3989|M.avium_104 -MVHSRMPALLTPGQTCWRTARADRFAA-----IIDAADYFRHVKAAMLR
Mflv_1004|M.gilvum_PYR-GCK ----MSVEGLLVPGQTCWQVATADRFAP-----IVDGADYLTHVKAAMLR
MSMEG_0375|M.smegmatis_MC2_155 MMAGLADWFLTAEERGNPHTSLPDWFTGNNVEVLVHGATYFDRLTDEVEK
* . : .* *: ::..* *: ::. : :
Mvan_5802|M.vanbaalenii_PYR-1 ARHRVMLIGWDFDTRMTFERGDKTLAGPNQLRAFLWWLLRLRPDLEIYLL
MAV_3989|M.avium_104 ARHRIMLVGWDFDSRVTFERGDKTLPGPNQLGAFLFWMVRKRPALQIHLL
Mflv_1004|M.gilvum_PYR-GCK AQRRIMLIGWDLDYRTTFEPAGATMSGPNQLGPFLHWLLWAHRDLKVYLL
MSMEG_0375|M.smegmatis_MC2_155 LGRGDHLFFTDWRG----DPDERTRDGGPTIAELFCRAAQRGVVVKGLMW
: *. * : * * : :: :: :
Mvan_5802|M.vanbaalenii_PYR-1 KSNLGLIPAFQTIWDGVTPVKLLNWLSSDRLWFAVDGAHPTGAVHHQKIV
MAV_3989|M.avium_104 KSNLRLLPALDGLWFGLTPVSLLNQISSRRMHFAIDGAHPVGSVHHQKIV
Mflv_1004|M.gilvum_PYR-GCK KSNLRLLPALDGFWFGVTPVALLNQITSRRMHFAVDGAHPAGAVHHQKIV
MSMEG_0375|M.smegmatis_MC2_155 RSHLDQFAYSE-----EQNRHLGEAIERAGGEVLLDQRVRIGGSHHQKLV
:*:* :. : * : : . :* *. ****:*
Mvan_5802|M.vanbaalenii_PYR-1 VVDD------VLAFCGGIDLTVDRWDTSEHLQHNTIRRTMGRAYG---PR
MAV_3989|M.avium_104 VVDD------AVAFCGGIDLTLDRWDTRAHEHTSRRRRTAGRGYG---PR
Mflv_1004|M.gilvum_PYR-GCK VIDD------AVAFCGGLDLTIGRWDTRAHEPEDPGRQTAGQPYG---PR
MSMEG_0375|M.smegmatis_MC2_155 VLRHPASPQRDVAFAGGIDLCHSRRDDAGHR-GDPQAVPMGRQYGDHPPW
*: . :**.**:** .* * * . . *: ** *
Mvan_5802|M.vanbaalenii_PYR-1 HEVAVAVDGAAARALGDQARNRWRAATKQLLGP-------------IEDA
MAV_3989|M.avium_104 HDVGVAVDGAAARALGEQARTRWQTATKQSLSP-------------VTAK
Mflv_1004|M.gilvum_PYR-GCK HEVAATVDGAAARVLSEQARNRWQAATGEALPG-------------LTAR
MSMEG_0375|M.smegmatis_MC2_155 HDAQLQIRGPAVGALDLTFRERWTDPAPLDMLSPIAWITDKLRRSDLSAD
*:. : *.*. .*. * ** .: : :
Mvan_5802|M.vanbaalenii_PYR-1 DRIWPPGLEPSLRRVGVGIARTLPAQT-----PGGEVREVEALNRAAIAA
MAV_3989|M.avium_104 HSAWPSKLRPTLHNVDVGIALTLPDLD-----DRPEVRQVEALNLAAIAA
Mflv_1004|M.gilvum_PYR-GCK ESVWPHTLQPALCDVEIGVARTLPALP-----GSEEVREVEALNLAAIAA
MSMEG_0375|M.smegmatis_MC2_155 PLPGRPPDPPPCGTQAVQVLRTYPDAHFEYDFAPRGERSIARGYTKAVLR
*. : : * * *.: *:
Mvan_5802|M.vanbaalenii_PYR-1 GRRTIYLENQYLASRPLVSALASRLQEPDGPEIVIVLPRR--SESRLERE
MAV_3989|M.avium_104 ARDTIYVENQYLASRTIAQALAQRLREEHGPQIVIVLARK--GNNPLERG
Mflv_1004|M.gilvum_PYR-GCK ARSVIYLENQYLASRSLVEALAARLREPDGPEIVIVLPRS--SESRLEQE
MSMEG_0375|M.smegmatis_MC2_155 ARRLIYLEDQYLWSKQVSRLFAAALAANPDLHLIAVVPRHPDVDGRLSLP
.* **:*:*** *: : :* * . .:: *:.* :. *.
Mvan_5802|M.vanbaalenii_PYR-1 SMDTARHDLLEILWAADDHHRLGVYWPVADGGAPVYVHSKVLVSDDRLLR
MAV_3989|M.avium_104 TMDSARHRLIRLLWDADEHHRLGVYWPVTDGGAPIYIHSKVLAVDDRLLR
Mflv_1004|M.gilvum_PYR-GCK SMDSARERLLRVLWEADEHDRLGVYWPAVAGGESVYVHAKVMVIDDRLLR
MSMEG_0375|M.smegmatis_MC2_155 PNQVGRQQAIATCRRAAP-DRVHVFDVENHKGTPVYVHAKVCVIDDVWAS
. : .*. : * .*: *: * .:*:*:** . **
Mvan_5802|M.vanbaalenii_PYR-1 IGSSNLNNRSMGFDSECDVAIEADPD--------DRDHDAVRRVITATRD
MAV_3989|M.avium_104 IGSSNFNNRSMGFDSECDVAIEAR----------GSQHDDVRREITSVRD
Mflv_1004|M.gilvum_PYR-GCK IGSSNLNNRSLGFDSECDLALESTP----------GGREDVRRLILHTRD
MSMEG_0375|M.smegmatis_MC2_155 VGSDNFNRRSWTHDSELSCAVLDDEREEREPRDPAGLGDGARVFARELRL
:**.*:*.** .*** . *: : .* *
Mvan_5802|M.vanbaalenii_PYR-1 GLVGEHLGVPAAEVASAVREHGSFLNALRALRGPGRTLVEFTADTVRDEA
MAV_3989|M.avium_104 ELVAEHLGVPVATLQEAIADRGSLLAAVEELRGDGKTLRPFTERTVANEA
Mflv_1004|M.gilvum_PYR-GCK DLVAEHLGVPTETFAREFARLGSFVAAVDALRGTGRSLRKFTEFMISADV
MSMEG_0375|M.smegmatis_MC2_155 KLLREHLDLDGKNDDAALIDPSDAVAAVNASANALQAWYD-SGRTGPRPP
*: ***.: . .. : *: . :: :
Mvan_5802|M.vanbaalenii_PYR-1 GRLAENDLMDPDRVPRSITRTVQRLFGRLTE-------------------
MAV_3989|M.avium_104 GPLAESELMDPDHVPRSLTRSVQRLITGRRG-------------------
Mflv_1004|M.gilvum_PYR-GCK SPFAENDLMDPDRVPRSITRSAWNVATTAITWPLMRTAPGLYSLIRAAAT
MSMEG_0375|M.smegmatis_MC2_155 GRLRPHRPERLGLLTRAWAEPVYRAVYDPDGRNYRDRLRRYW--------
. : . :.*: :... .
Mvan_5802|M.vanbaalenii_PYR-1 --
MAV_3989|M.avium_104 --
Mflv_1004|M.gilvum_PYR-GCK RS
MSMEG_0375|M.smegmatis_MC2_155 --