For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLYDTLSGGVRDFAPLRPGHVSIYLCGATVQGLPHIGHVRSGVAFDVLRRWLTAKGFDVAFIRNVTDIDD KILHKAADAGRPWWEWAATYERAFTAAYDALGVLPPSAEPRATGHITQMVELIERLIERGHAYTGDGDVY FSVATLPDYGKLSGHRIDDVHQGEGVATGKRDQRDFTLWKGAKPGEPSWPTPWGRGRPGWHTECVAMCES YLGAAFDIHAGGMDLVFPHHENEIAQAEAAGDGFARFWLHNGWVTMGGEKMSKSLGNVLAIPAVLQRVRA AELRYYLGSAHYRSMLEFSETALQDAAKAYAGIEDFLHRVRTRVGAVVPGEWTPKFGAALDDDLAVPAAL AEVHAARAEGNRALDAGDHDTALTHAMSIRAMMGILGADPLDERWESRDETSAALAAVDVLVQAEVQRRE DARARRDWAEADAIRDRLKEAGIEVTDTADGPQWSLLDGTDK*
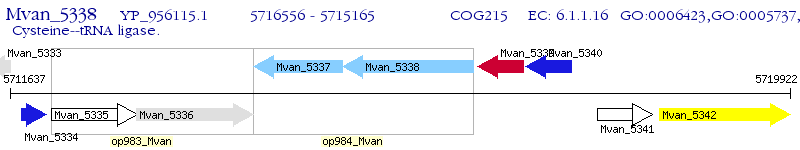
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5338 | cysS | - | 100% (463) | cysteinyl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_3478 | - | 2e-57 | 35.64% (404) | cysteinyl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3611c | cysS | 0.0 | 83.15% (463) | cysteinyl-tRNA synthetase |
| M. gilvum PYR-GCK | Mflv_1446 | cysS | 0.0 | 95.07% (467) | cysteinyl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv3580c | cysS | 0.0 | 83.15% (463) | cysteinyl-tRNA synthetase |
| M. leprae Br4923 | MLBr_00323 | cysS | 0.0 | 81.74% (460) | cysteinyl-tRNA synthetase |
| M. abscessus ATCC 19977 | MAB_0571 | cysS | 0.0 | 79.66% (467) | cysteinyl-tRNA synthetase |
| M. marinum M | MMAR_5080 | cysS | 0.0 | 83.15% (463) | cysteinyl-tRNA synthetase 1 CysS1 |
| M. avium 104 | MAV_0573 | cysS | 0.0 | 84.90% (457) | cysteinyl-tRNA synthetase |
| M. smegmatis MC2 155 | MSMEG_6074 | cysS | 0.0 | 86.49% (459) | cysteinyl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_3755 | - | 0.0 | 84.73% (465) | PUTATIVE cysteinyl-tRNA synthetase |
| M. ulcerans Agy99 | MUL_4156 | cysS | 0.0 | 83.15% (463) | cysteinyl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5338|M.vanbaalenii_PYR-1 ---------------MRLYDTLSGGVRDFAPLRPGHVSIYLCGATVQGLP
Mflv_1446|M.gilvum_PYR-GCK ---------------MRLYDTLSGGVRDFAPLRPGHVSIYLCGATVQGLP
MSMEG_6074|M.smegmatis_MC2_155 MTDRAQAVGSGPELGLRLYDTMAGAVRDFVPLRPGHASIYLCGATVQGLP
TH_3755|M.thermoresistible__bu --------VTNSPAGLRLHDTMAGAVREFVPVRAGHVSIYLCGATVQGLP
Mb3611c|M.bovis_AF2122/97 ---------MTDRARLRLHDTAAGVVRDFVPLRPGHVSIYLCGATVQGLP
Rv3580c|M.tuberculosis_H37Rv ---------MTDRARLRLHDTAAGVVRDFVPLRPGHVSIYLCGATVQGLP
MLBr_00323|M.leprae_Br4923 ---------MIDSGHLRLHDTVAGAVRDFVPLRAGHVSIYLCGATVQGQP
MMAR_5080|M.marinum_M ---------MTERAGLRLHDTAAGAVRDFVPLREGHVSIYLCGATVQGLP
MUL_4156|M.ulcerans_Agy99 ---------MTERAGLRLHDTAAGAVRDFVPLREGHVSIYLCGATVQGLP
MAV_0573|M.avium_104 ---------MTDRPRLRLHDTAAGAVRDFVPLRDGHVSIYLCGATVQGLP
MAB_0571|M.abscessus_ATCC_1997 ---------MTGHVPLRLYDTRVGAVRDFVPLQPGTVSIYLCGATVQGLP
:**:** * **:*.*:: * .*********** *
Mvan_5338|M.vanbaalenii_PYR-1 HIGHVRSGVAFDVLRRWLTAKGFDVAFIRNVTDIDDKILHKAADAGRPWW
Mflv_1446|M.gilvum_PYR-GCK HIGHVRSGVAFDVLRRWLTAKGFDVAFIRNVTDIDDKILIKAADAGRPWW
MSMEG_6074|M.smegmatis_MC2_155 HIGHVRSGVAFDVLRRWLTANGYDVAFIRNVTDIDDKILNKAADAGRPWW
TH_3755|M.thermoresistible__bu HIGHVRSGVAFDVLRRWLMAKGYDVAFIRNVTDIDDKILAKAAEAGRPWW
Mb3611c|M.bovis_AF2122/97 HIGHVRSGVAFDILRRWLLARGYDVAFIRNVTDIEDKILAKAAAAGRPWW
Rv3580c|M.tuberculosis_H37Rv HIGHVRSGVAFDILRRWLLARGYDVAFIRNVTDIEDKILAKAAAAGRPWW
MLBr_00323|M.leprae_Br4923 HIGHVRSGVAFDILRRWLMARGYDVAFIRNVTDIDDKILNKAAAAGRPWW
MMAR_5080|M.marinum_M HIGHVRSGVAFDILRRWLMARGFDVAFIRNVTDIDDKILNKAAAAERPWW
MUL_4156|M.ulcerans_Agy99 HIGHVRSGVAFDILRRWLMARGFDVAFIRNVTDIDDKILNKAAAAERPWW
MAV_0573|M.avium_104 HIGHVRSGVAFDILRRWLIALGYDVAFIRNVTDIDDKILNKAAAAGRPWW
MAB_0571|M.abscessus_ATCC_1997 HIGHVRSGVAFDVLRRWLSARAYDVLFVRNVTDIDDKILNKAADAGRPWW
************:***** * .:** *:******:**** *** * ****
Mvan_5338|M.vanbaalenii_PYR-1 EWAATYERAFTAAYDALGVLPPSAEPRATGHITQMVELIERLIERGHAYT
Mflv_1446|M.gilvum_PYR-GCK EWAATYERAFTAAYDALGVLPPSAEPRATGHITQMVELIDRLIERGHAYC
MSMEG_6074|M.smegmatis_MC2_155 EWAATFERAFSAAYDALGVLPPSAEPRATGHITQMVELIERLIDKGHAYA
TH_3755|M.thermoresistible__bu EWAATYERAFTAAYDALGVLPPSAEPRATGHITQMVELIERLLEKGHAYR
Mb3611c|M.bovis_AF2122/97 EWAATHERAFTAAYDALDVLPPSAEPRATGHITQMIEMIERLIQAGHAYT
Rv3580c|M.tuberculosis_H37Rv EWAATHERAFTAAYDALDVLPPSAEPRATGHITQMIEMIERLIQAGHAYT
MLBr_00323|M.leprae_Br4923 EWAATHERAFTAAYDALDVLPPSAEPRATGHITQMIELIELLIETGHAYT
MMAR_5080|M.marinum_M EWAATYERAFTAAYDALDVLPPSAEPRATGHVTQMVELIERLIERGHAYA
MUL_4156|M.ulcerans_Agy99 EWAATYERAFTAAYDALDVLPPSAEPRATGHVTQMVELIERLIERGHAYA
MAV_0573|M.avium_104 EWAATYERAFSAAYDALDVLPPSAEPRATGHITQMVELIERLIEKGHAYT
MAB_0571|M.abscessus_ATCC_1997 EWAATYERAFDEAYESLGVLPPSVSPRATGHITQMIELIDRLIERGHAYR
*****.**** **::*.*****..******:***:*:*: *:: ****
Mvan_5338|M.vanbaalenii_PYR-1 GD----GDVYFSVATLPDYGKLSGHRIDDVHQGEGVATGKRDQRDFTLWK
Mflv_1446|M.gilvum_PYR-GCK ADQEGNGDVYFSVSTLPEYGKLSGHRIDDVHQGEGVATGKRDQRDFTLWK
MSMEG_6074|M.smegmatis_MC2_155 AG----GDVYFDVLSLPDYGQLSGHRIDDVHQGEGVATGKRDQRDFTLWK
TH_3755|M.thermoresistible__bu ANPDGDGDVYFDVLSFPEYGRLSGHRIEDVHQGEGVATGKRDQRDFTLWK
Mb3611c|M.bovis_AF2122/97 GG----GDVYFDVLSYPEYGQLSGHKIDDVHQGEGVAAGKRDQRDFTLWK
Rv3580c|M.tuberculosis_H37Rv GG----GDVYFDVLSYPEYGQLSGHKIDDVHQGEGVAAGKRDQRDFTLWK
MLBr_00323|M.leprae_Br4923 GG----SDVYFDVLSYPDYGQLSGHKMDYIHQGEGVTTGKRDQRDFTLWK
MMAR_5080|M.marinum_M GG----GDVYFDVLSYPDYGQLSGHKVDDVHQGEGVATGKRDQRDFTLWK
MUL_4156|M.ulcerans_Agy99 GG----GDVYFDVLSYPDYGQLSGHKVDDVHQGEGVATGKRDQRDFTLWK
MAV_0573|M.avium_104 GD----GDVYFDVLSYPEYGQLSGHRIDDVHQGEGVASGKRDQRDFTLWK
MAB_0571|M.abscessus_ATCC_1997 DLREGGGDVYFDVLSYPEYGQLSGHKIDDVHQGEGVATGKRDPRDFTLWK
.****.* : *:**:****::: :******::**** *******
Mvan_5338|M.vanbaalenii_PYR-1 GAKPGEPSWPTPWGRGRPGWHTECVAMCESYLGAAFDIHAGGMDLVFPHH
Mflv_1446|M.gilvum_PYR-GCK GAKPGEPSWPTPWGRGRPGWHTECVAMCEAYLGAEFDIHAGGMDLVFPHH
MSMEG_6074|M.smegmatis_MC2_155 GAKPGEPSWPTPWGRGRPGWHTECVAMCEAYLGPEFDIHAGGMDLVFPHH
TH_3755|M.thermoresistible__bu SAKPGEPSWPTPWGRGRPGWHLECSAMAHTYLGSEFDIHCGGMDLVFPHH
Mb3611c|M.bovis_AF2122/97 GEKPGEPSWPTPWGRGRPGWHLECSAMARSYLGPEFDIHCGGMDLVFPHH
Rv3580c|M.tuberculosis_H37Rv GEKPGEPSWPTPWGRGRPGWHLECSAMARSYLGPEFDIHCGGMDLVFPHH
MLBr_00323|M.leprae_Br4923 GAKSGEPSWPTPWGRGRPGWHLECSAMARAYLGSEFDIHCGGMDLVFPHH
MMAR_5080|M.marinum_M GAKPGEPSWPTPWGRGRPGWHLECSAMARTYLGAQFDIHCGGMDLIFPHH
MUL_4156|M.ulcerans_Agy99 GAKPGEPSWPTPWGRGRPGWHLECSAMARTYLGAQFDIHCGGMDLIFPHH
MAV_0573|M.avium_104 GAKPGEPSWPTPWGRGRPGWHTECVAMAHEYLGPEFDIHCGGMDLVFPHH
MAB_0571|M.abscessus_ATCC_1997 GAKPGEPSWPTPWGPGRPGWHLECSAMARTYLGPSFDIHCGGMDLVFPHH
. *.********** ****** ** **.. ***. ****.*****:****
Mvan_5338|M.vanbaalenii_PYR-1 ENEIAQAEAAGDGFARFWLHNGWVTMGGEKMSKSLGNVLAIPAVLQRVRA
Mflv_1446|M.gilvum_PYR-GCK ENEIAQAEAAGDGFARFWLHNGWVTMGGEKMSKSLGNVLAIPAVLQRVRA
MSMEG_6074|M.smegmatis_MC2_155 ENEIAQAHGAGDGFARYWLHNGWVTMGGEKMSKSLGNVLSIPAVLQRVRA
TH_3755|M.thermoresistible__bu ENEIAQSRAAGDGFAQYWLHNGWVTMGGEKMSKSLGNVLAIPAVLERVRA
Mb3611c|M.bovis_AF2122/97 ENEIAQSRAAGDGFARYWLHNGWVTMGGEKMSKSLGNVLSMPAMLQRVRP
Rv3580c|M.tuberculosis_H37Rv ENEIAQSRAAGDGFARYWLHNGWVTMGGEKMSKSLGNVLSMPAMLQRVRP
MLBr_00323|M.leprae_Br4923 ENEIAQSRAVGDGFARYWLHNGWVTMGGEKMSKSLGNVLSIPAVLQRVRP
MMAR_5080|M.marinum_M ENEIAQSRAAGDGFARYWLHNGWVTMGGEKMSKSLGNVLAIPTMLQRVRP
MUL_4156|M.ulcerans_Agy99 ENEIAQSRAAGDGFARYWLHNGWVTMGGEKMSKSLGNVLAIPTMLQRVRP
MAV_0573|M.avium_104 ENEIAQSRAAGDGFARYWLHNGWVTMGGEKMSKSLGNVLAIPAMLQRVRP
MAB_0571|M.abscessus_ATCC_1997 ENEIAQSCAAGDGFARYWMHNGWVTMGGEKMSKSLGNVLSIPAVLQRVRA
******: ..*****::*:********************::*::*:***.
Mvan_5338|M.vanbaalenii_PYR-1 AELRYYLGSAHYRSMLEFSETALQDAAKAYAGIEDFLHRVRTRVGAVVPG
Mflv_1446|M.gilvum_PYR-GCK AELRYYLGSAHYRSMLEFSENALQDAARAYSGVEDFLHRVRVRVGAVVPG
MSMEG_6074|M.smegmatis_MC2_155 AELRYYLGSAHYRSMLEFSETALQDAVKAYTGIEDFLHRVCSRVGSVPIG
TH_3755|M.thermoresistible__bu AELRYYLGSAHYRSMLEFSETALQDAAKAYRGIEDFLHRVRIRVGGVEPG
Mb3611c|M.bovis_AF2122/97 AELRYYLGSAHYRSMLEFSETAMQDAVKAYVGLEDFLHRVRTRVGAVCPG
Rv3580c|M.tuberculosis_H37Rv AELRYYLGSAHYRSMLEFSETAMQDAVKAYVGLEDFLHRVRTRVGAVCPG
MLBr_00323|M.leprae_Br4923 AELRYYLGSAHYRSMLEFSEAALQDAVKAYVGVENFLTRVRTRVGAVGTG
MMAR_5080|M.marinum_M AELRYYLGSAHYRSMLEFSDIALQDAVNAYVGVEEFLHRVRSRAGVVVVG
MUL_4156|M.ulcerans_Agy99 AELRYYLGSAHYRSILEFSDIALQDAVNAYVGVEEFLHRVRSRVGAVVVG
MAV_0573|M.avium_104 AELRYYLGSAHYRSMLEFSDTALQDAVKAYVGVEEFLHRVRVRVGAVEPG
MAB_0571|M.abscessus_ATCC_1997 VELRYYLGSAHYRSMLEYSETALADAVKAYCGIEDFLHRVRIRVGDVPVG
.*************:**:*: *: **..** *:*:** ** *.* * *
Mvan_5338|M.vanbaalenii_PYR-1 EWTPKFGAALDDDLAVPAALAEVHAARAEGNRALDAGDHDTALTHAMSIR
Mflv_1446|M.gilvum_PYR-GCK EWTPKFGAALDDDLAVPAALAEVHAARAEGNRALDAGDHDAALTYAMSIR
MSMEG_6074|M.smegmatis_MC2_155 EWTPKFAAALNDDLSVPIALAEIHAARAEGNRALDSGDHQTAMQQASSIR
TH_3755|M.thermoresistible__bu EWTPKFAAALDDDLSVPIALAEVHHARAEGNRALNAGDHESALEHARSIR
Mb3611c|M.bovis_AF2122/97 DPTPRFAEALDDDLSVPIALAEIHHVRAEGNRALDAGDHDGALRSASAIR
Rv3580c|M.tuberculosis_H37Rv DPTPRFAEALDDDLSVPIALAEIHHVRAEGNRALDAGDHDGALRSASAIR
MLBr_00323|M.leprae_Br4923 ELTPRFAAALDDDLAVPIALAEVHHARVEGNRALDIGDHEGALTNAGAIR
MMAR_5080|M.marinum_M DWTPRFAAALDDDLSVPIALAEIHHTRAEGNRALDAGDHDAALQSASSIR
MUL_4156|M.ulcerans_Agy99 DWTPRFAAALDDDLSVPIALAEIHHIRAEGNRALDAGDHDAALQSASSIR
MAV_0573|M.avium_104 EPTPRFADALNDDLAVPAALAEVHQARAEGNRALDSGDHEGALRQARSIR
MAB_0571|M.abscessus_ATCC_1997 TWTPAFAAALDDDLSVPIALAEVHSTRADGNRALDAGDHEGALSAAASIR
** *. **:***:** ****:* *.:*****: ***: *: * :**
Mvan_5338|M.vanbaalenii_PYR-1 AMMGILGADPLDERWESRDETSAALAAVDVLVQAEVQRREDARARRDWAE
Mflv_1446|M.gilvum_PYR-GCK AMMGILGADPLDERWESRDKTSAALAAVDVLVHAEVRRREDARARRDWAE
MSMEG_6074|M.smegmatis_MC2_155 AMMDILGCDPLNERWESRDATSAALKAVDVLVQWALASRAEARERRDWAA
TH_3755|M.thermoresistible__bu AMMGILGCDPLDERWETDRESSAALAALDVLVRWALEEREEARARRDWAQ
Mb3611c|M.bovis_AF2122/97 AMMGILGCDPLDQRWESRDETSAALAAVDVLVQAELQNREKAREQRNWAL
Rv3580c|M.tuberculosis_H37Rv AMMGILGCDPLDQRWESRDETSAALAAVDVLVQAELQNREKAREQRNWAL
MLBr_00323|M.leprae_Br4923 AMMGILGCDPLDERWESRDETSAALAAVDVLVAAELESRQMAREQRNWVL
MMAR_5080|M.marinum_M AMMGILGCDPLDERWESRDETSAALAAVDVLVQAELENRQKAREERNWAL
MUL_4156|M.ulcerans_Agy99 ALMGILGCDPLDERWESRDETSAALAAVDVLVQAELENRQKAREERNWAL
MAV_0573|M.avium_104 AMMGILGCDPLHERWETRDESSAALAAVDVLVRAELQNREKAREQRNWAL
MAB_0571|M.abscessus_ATCC_1997 AMMSILGCDPLDERWETRDETSAALNAVDVLVTAELQRRETARTEKNWAL
*:*.***.***.:***: :**** *:**** : * ** .::*.
Mvan_5338|M.vanbaalenii_PYR-1 ADAIRDRLKEAGIEVTDTADGPQWSLLDGTDK------------------
Mflv_1446|M.gilvum_PYR-GCK ADAIRDRLKEAGIEVTDTGDGPQWSLLDGTDK------------------
MSMEG_6074|M.smegmatis_MC2_155 ADAIRDRLKEAGIEVTDTADGPQWALIE-RDK------------------
TH_3755|M.thermoresistible__bu ADTIRDRLKEAGIEVTDTPDGPQWSLLDVTAVMXXXTSPYRAGPARRAAA
Mb3611c|M.bovis_AF2122/97 ADEIRGRLKRAGIEVTDTADGPQWSLLGGDTK------------------
Rv3580c|M.tuberculosis_H37Rv ADEIRGRLKRAGIEVTDTADGPQWSLLGGDTK------------------
MLBr_00323|M.leprae_Br4923 ADQIRDRLKDAGIEVTDTVNGPQWELLAGDKQVDAR--------------
MMAR_5080|M.marinum_M ADEIRNRLKNAGIEVTDTADGPQW-LLGGDGK------------------
MUL_4156|M.ulcerans_Agy99 ADEIRNRLKNAGIEVTDTADGPQW-LLGGDGK------------------
MAV_0573|M.avium_104 ADEIRNRLKQAGIEVTDTADGPQW-TLGGDGK------------------
MAB_0571|M.abscessus_ATCC_1997 ADEIRDRLKRAGLEITDTADGPQWTLTSSDEEQYHHGR------------
** **.*** **:*:*** :****
Mvan_5338|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1446|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6074|M.smegmatis_MC2_155 --------------------------------------------------
TH_3755|M.thermoresistible__bu AHLLPAAAARSRRAPGGVGRADPQPARRAGRQRPRLLAGHPRHRRGAARR
Mb3611c|M.bovis_AF2122/97 --------------------------------------------------
Rv3580c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00323|M.leprae_Br4923 --------------------------------------------------
MMAR_5080|M.marinum_M --------------------------------------------------
MUL_4156|M.ulcerans_Agy99 --------------------------------------------------
MAV_0573|M.avium_104 --------------------------------------------------
MAB_0571|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5338|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1446|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6074|M.smegmatis_MC2_155 --------------------------------------------------
TH_3755|M.thermoresistible__bu IRRQGRTGAVAADRPADPAPARHPHRPAQSAPAAPGRRRGRRAVARRPDR
Mb3611c|M.bovis_AF2122/97 --------------------------------------------------
Rv3580c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00323|M.leprae_Br4923 --------------------------------------------------
MMAR_5080|M.marinum_M --------------------------------------------------
MUL_4156|M.ulcerans_Agy99 --------------------------------------------------
MAV_0573|M.avium_104 --------------------------------------------------
MAB_0571|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5338|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1446|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6074|M.smegmatis_MC2_155 --------------------------------------------------
TH_3755|M.thermoresistible__bu GRLPGGHHLVRPGPRTVPAGAVPGRGRRRPAGAAATVELPPPRRPALDGA
Mb3611c|M.bovis_AF2122/97 --------------------------------------------------
Rv3580c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00323|M.leprae_Br4923 --------------------------------------------------
MMAR_5080|M.marinum_M --------------------------------------------------
MUL_4156|M.ulcerans_Agy99 --------------------------------------------------
MAV_0573|M.avium_104 --------------------------------------------------
MAB_0571|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5338|M.vanbaalenii_PYR-1 --------------------------------------
Mflv_1446|M.gilvum_PYR-GCK --------------------------------------
MSMEG_6074|M.smegmatis_MC2_155 --------------------------------------
TH_3755|M.thermoresistible__bu RLRRRRHPAPLPAAGHRRPSDGVPGAGRRAAVTHRRPS
Mb3611c|M.bovis_AF2122/97 --------------------------------------
Rv3580c|M.tuberculosis_H37Rv --------------------------------------
MLBr_00323|M.leprae_Br4923 --------------------------------------
MMAR_5080|M.marinum_M --------------------------------------
MUL_4156|M.ulcerans_Agy99 --------------------------------------
MAV_0573|M.avium_104 --------------------------------------
MAB_0571|M.abscessus_ATCC_1997 --------------------------------------