For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNPRWLYRVGVLSCAAALTACSSGGDNAESAGTSEPTSPVQSAPIPEPLVATYDGGLYVLDGETLELRQD IPLDGFLRVNPAGDDAHVLVTTNEGFRILDAASGQLTDETFPAAEAGHVVPHGETTVLFADGSGEITAFD PHALADGMPETQTFTTPQPHHGVAVILSDGTLVHSLGDAESRTGAVALQGDREIARNEDCPGLHGETVVA DEEVVLGCENGALIFANGRFTKVASPTPYGRIGNIKGHDDSPVALGDMKIDPDAELERPTQFSLIDTTTD KLTLVQLPPSVSYSFRSLARGPQAEALILGTDGKLYVFDPVTGAQIKTIDVTGAWTEPDDWQDPRPTVFT REDVVYVTDPATRQIHLLDLQSGTVTASVTLDQAPNELSGAVGHDH*
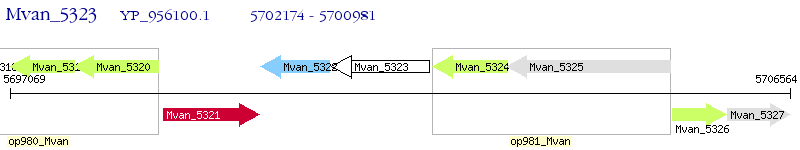
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5323 | - | - | 100% (397) | hypothetical protein Mvan_5323 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3560 | - | 1e-91 | 44.67% (403) | putative secreted protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6049 | - | 1e-153 | 67.10% (389) | secreted protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5323|M.vanbaalenii_PYR-1 MSNPRWLYRVGVLSCAAALTACSSGGDNAESAGTSEPTSPVQSAPIPEPL
MSMEG_6049|M.smegmatis_MC2_155 MARP--------LPLTAALGALALISTLAGCSGEAERST---AEPIAEPL
Mflv_3560|M.gilvum_PYR-GCK MTTTTRLFTFFSVALLGAGCAATETAPTDSAAPASAEAAPTEESSATPRL
*: . :. .* * : . .: : :: . . *
Mvan_5323|M.vanbaalenii_PYR-1 VATYDGGLYVLDGETLELRQDIPLDGFLRVNPAGDDAHVLVTTNEGFRIL
MSMEG_6049|M.smegmatis_MC2_155 VATYDGGLYVLDGTTLDVVADIPMEGFLRVNPAGDDAHVLVTTNEGFRLL
Mflv_3560|M.gilvum_PYR-GCK ALSYDGGVLVLDAGTLHQVADIPVDGFVRLNSAADGRHVLVSQTDGFAVL
. :****: ***. **. ***::**:*:*.*.*. ****: .:** :*
Mvan_5323|M.vanbaalenii_PYR-1 DA---------------ASGQLTDETFPAAEAGHVVPHGETTVLFADGSG
MSMEG_6049|M.smegmatis_MC2_155 DA---------------KSGELTETTFTASEPGHVVPHGERTALFADGSG
Mflv_3560|M.gilvum_PYR-GCK DLGTWTDSHGDHGHHYTAPPAMTDLRFGGAEPGHVVAHDERLTLFSDGTG
* . :*: * .:*.****.*.* .**:**:*
Mvan_5323|M.vanbaalenii_PYR-1 EITAFDPHALADGMPETQTFTTPQPHHGVAVILSDGTLVHSLGDAESRTG
MSMEG_6049|M.smegmatis_MC2_155 EITIFDPHELADGKPEVTTLSSPKPHHGVAVVLSDGSFIRTEGDPDERSG
Mflv_3560|M.gilvum_PYR-GCK AVDVVDPHELLDGEGVATTVVTTTPHHGVAVARKDGTVVLSVGDEDGRSG
: .*** * ** . *. :. ******* .**:.: : ** : *:*
Mvan_5323|M.vanbaalenii_PYR-1 AVALQGD-REIARNEDCPGLHGETVVADEEVVLGCENGALIFANGRFTKV
MSMEG_6049|M.smegmatis_MC2_155 AVAFDSNQAEIARSAECPGIHGETVVEGETVVFGCENGALTYSDRVFTKI
Mflv_3560|M.gilvum_PYR-GCK LEIRDAAGGRIAVNDQCPGLHGEAAAAGGALTFGCEDGLLIVRGNDIQKI
:. .** . :***:***:.. . :.:***:* * . : *:
Mvan_5323|M.vanbaalenii_PYR-1 ASPTPYGRIGNIKGHDDSPVALGDMKIDPDAELERPTQFSLIDTTTDKLT
MSMEG_6049|M.smegmatis_MC2_155 AAPGDYGRIGTVKGHDDSAVALGDFKVDPDAELERPNQFALIDTAGNRLQ
Mflv_3560|M.gilvum_PYR-GCK PSPDPYGRIGNQAGSEESVVVLGDYKTDPDAELERPQRFTLTDTRTGALR
.:* *****. * ::* *.*** * ********* :*:* ** . *
Mvan_5323|M.vanbaalenii_PYR-1 LVQLPPSVSYSFRSLARGPQAEALILGTDGKLYVFDPVTGAQIKTIDVTG
MSMEG_6049|M.smegmatis_MC2_155 LVTLPPSVSYSFRSLARGPHAEALILGTDGKLYVIDPASGDTVKTIAVTE
Mflv_3560|M.gilvum_PYR-GCK VVPLD--ASYSFRSLDRGPRGEAVILGTDGALHVFDPVSAQRLAHVPVVE
:* * .******* ***:.**:****** *:*:**.:. : : *.
Mvan_5323|M.vanbaalenii_PYR-1 AWTEPDDWQDPRPTVFTREDVVYVTDPATRQIHLLDLQSGTVTASVTLDQ
MSMEG_6049|M.smegmatis_MC2_155 PWTEPDDWQQPRPTVFTRDHDVYVTDPATRQLHLVDIESGAVTKTATLEN
Mflv_3560|M.gilvum_PYR-GCK PWTEPDEWQSPMPNLHVQGDVAYVSDPGTRRVVAVNLSDGAVLAETTLAH
.*****:**.* *.:..: . .**:**.**:: :::..*:* .** :
Mvan_5323|M.vanbaalenii_PYR-1 APNELSGAVGHDH-
MSMEG_6049|M.smegmatis_MC2_155 APNELSGAVGHEHS
Mflv_3560|M.gilvum_PYR-GCK PTIELTGVEG----
.. **:*. *