For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARLPYFAALTLAGGLIASAVTAPTAFTQPPAPVAGSESPGDTIRELTNQGFDVQINWVAGQPSNIPLSQC TVTRIDTSAPPTAWVSINCPPDGSQ*
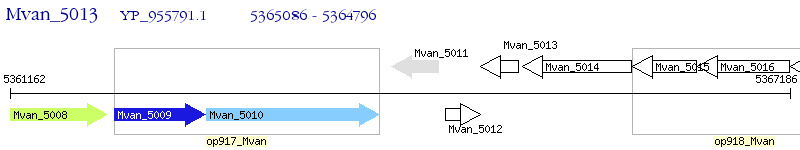
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5013 | - | - | 100% (96) | hypothetical protein Mvan_5013 |
| M. vanbaalenii PYR-1 | Mvan_4705 | - | 1e-04 | 35.53% (76) | hypothetical protein Mvan_4705 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1738 | - | 1e-33 | 64.58% (96) | hypothetical protein Mflv_1738 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1850c | - | 7e-11 | 40.22% (92) | hypothetical protein MAB_1850c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5321 | - | 8e-05 | 37.97% (79) | hypothetical protein MSMEG_5321 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5013|M.vanbaalenii_PYR-1 MARLP-YFAALTLAGGLIA-SAVTAPTAFTQPPAPVAGSESPGDTIRELT
Mflv_1738|M.gilvum_PYR-GCK MMRTM-RFAGVPAAAAVIG-SLAVVAPATAQPSLPEAGNETPGATIRQLS
MAB_1850c|M.abscessus_ATCC_199 MASATGRIALMPLWLGAIALSLAIPAVAAAAPALPVGG-ESPSQTISDLE
MSMEG_5321|M.smegmatis_MC2_155 MTSTRTRQAFAAAGAGAAA---LSALLAFATP----AHAESAALTIGQLE
* * . . . * : * . *:.. ** :*
Mvan_5013|M.vanbaalenii_PYR-1 NQGFDVQINWVAGQPSNIPLSQCTVTRIDT-------------SAPP---
Mflv_1738|M.gilvum_PYR-GCK NAGYDVEVNWVAGEPSNIPLSQCTVTRIDT-------------SAPP---
MAB_1850c|M.abscessus_ATCC_199 DQGYDVQINYVMGN-SNTPLDMCRVLAVHNPNR----------SGKPVDS
MSMEG_5321|M.smegmatis_MC2_155 AQGFDVRVDRVGSA----PLEQCVVTGIRNPRERTRIVRFDGPRGRDRLV
*:**.:: * . **. * * : . .
Mvan_5013|M.vanbaalenii_PYR-1 --TAWVSINCPPDGSQ--
Mflv_1738|M.gilvum_PYR-GCK --TAWVSVNCPPDGSQ--
MAB_1850c|M.abscessus_ATCC_199 FTTVYVDISCP-DFWQDD
MSMEG_5321|M.smegmatis_MC2_155 PVVVQRSITVSLDCSRR-
.. .:. . * :