For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DSRTVRRRVLAATAVLAATLVPSCTRVVDDARAVAAVPGAPFGSYASADACTPVDSELADVPPPRGAAAD IEPVLRIPRPEGWKRVTALDSGAIRYVMRNEGLATGGITPTAVVTLESHRGELSPREFFADQRRQVMQLG VTDLTLTDTTLCGLPAVRGHYMSPLLGTAAPRRADLLSAVMVSEGTTYGVAVTVQSADERDPEYQRDSAQ ILDGFQMLPPGSR*
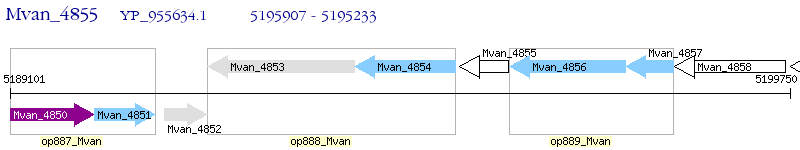
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4855 | - | - | 100% (224) | hypothetical protein Mvan_4855 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1876 | - | 5e-84 | 69.59% (217) | hypothetical protein Mflv_1876 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1893 | - | 9e-21 | 33.18% (211) | hypothetical protein MAB_1893 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5514 | - | 3e-41 | 44.65% (215) | hypothetical protein MSMEG_5514 |
| M. thermoresistible (build 8) | TH_1333 | - | 5e-38 | 44.29% (219) | hypothetical protein MSMEG_5514 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4855|M.vanbaalenii_PYR-1 --------------MDSRTVRRRVLAATAVLAATLVPSCTRVVDDARAVA
Mflv_1876|M.gilvum_PYR-GCK --------------MTRWS--RGAVAAVAAVTMALVPSCTHVVQDARGVA
MSMEG_5514|M.smegmatis_MC2_155 ------------------MTTAPKAAAVLFALTLLTASCTRVVDGRSVAG
TH_1333|M.thermoresistible__bu MFREQSQNRPETRGIGRFRVRSAGVAAVVALVTLLTVSCTRYVDAEPVAA
MAB_1893|M.abscessus_ATCC_1997 -----------------MIRPPRWMLAAVCAATMLTSGCTAVVDGAAVAT
*. *. .** *: .
Mvan_4855|M.vanbaalenii_PYR-1 AVPGAPFGSYAS-ADACTPVDSELADVPPPRGAAADIEPVLRIPRPEGWK
Mflv_1876|M.gilvum_PYR-GCK ARPGQVFGTAAS-VDECTWVEGELVDVPTPRGVSAQTDPVLRIPQPEGWN
MSMEG_5514|M.smegmatis_MC2_155 PDLAGGAASGAS-AAECEEVDAPLTDID----TQAAGEPILRIPQPQGWT
TH_1333|M.thermoresistible__bu EPVAADDRPAASDASPCTPVDPPLTAIP----ARADGEPVLRIPQPDGWE
MAB_1893|M.abscessus_ATCC_1997 PGEEGKWLTNPK----CSSVSVPLLDVP----LDNDSEPRVEVPQPTGWE
. .. * *. * : :* :.:*:* **
Mvan_4855|M.vanbaalenii_PYR-1 RVTALDSGAIRYVMRNEGLATGGITPTAVVTLESHRGELSPREFFADQRR
Mflv_1876|M.gilvum_PYR-GCK RITVLDSQMIRFVMRADDLAVRGVAPTAVVTLESHRGEISARDFFDEQRK
MSMEG_5514|M.smegmatis_MC2_155 RNTMLDSELIRFAMSNPALGTEEFAPTAVVTLETARGEQDPELVFENQRE
TH_1333|M.thermoresistible__bu RSRQLDSELIRFVIGNPGIVAEEFVPNVVVTLETVDG-VDPGLAFEQQRL
MAB_1893|M.abscessus_ATCC_1997 RINRFESGVVRVYLAAPDLQAGGFVPNATVAIANLSGKAGTEEAAFAAER
* ::* :* : : . ..*...*:: . * .. .
Mvan_4855|M.vanbaalenii_PYR-1 QVMQ-LGVTDLTLTDTTLCGLPAVRGHYMSPLLGTAAPRRADLLSAVMVS
Mflv_1876|M.gilvum_PYR-GCK QVSD-LGVKELTLEDTTLCGLPAVRGHYMAPLLGTSSPRRADLLSAVMYT
MSMEG_5514|M.smegmatis_MC2_155 ALETGLGATNISSEKHTLCGLPAETIHYQMPQMGGLEPHPAIVVGAVLHT
TH_1333|M.thermoresistible__bu GLEQVVGATDLRVSAHTLCGLPAQTIDYLTPAMGAVRPHPATVLLVALDT
MAB_1893|M.abscessus_ATCC_1997 GGLESFGVTDLVEAQGAICGYPSRTMTYNMG-LGNIPAHRVTTTIVAVKN
.*..:: ::** *: * :* .: . ..: .
Mvan_4855|M.vanbaalenii_PYR-1 EGTTYGVAVTVQSADERDPEYQRDSAQILDGFQMLPPGSR
Mflv_1876|M.gilvum_PYR-GCK EGRTFGVAVTVQSADEDDPVYQRDKAAILDGFQMLPPGTR
MSMEG_5514|M.smegmatis_MC2_155 ETKTFVAAVTVQTTDPDNPAYQRDAETIIKGFQILPPGER
TH_1333|M.thermoresistible__bu DDEMYIVAVTAQTMDPDNPTYRRDVDAILSGFVVLPPSPE
MAB_1893|M.abscessus_ATCC_1997 NTKMYTVGVSVQALDDTVRGFDTARETILAGLQVGPPATA
: : ..*:.*: * : *: *: : **.