For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFARSRGKGQPPAVVDEDPGAAAKVLSQIIERGARVQGPAVKAYVERLRDNNPGATPAEIIDKLDKKYLA AVMASGAAVGSTAAYPGIGTLVAMSAVAGETVVFLEATALYVLAVAEVHGIPAEHRERRRALVLSVLVGE DSKRAVADLLGSGRTSGAWVSDGAATLPLPALSQLNSRLLRYFVKRYTLKRGALAFGKMLPVGIGAVVGG VGNRFMGKKIIANAHNAFGAAPARWPSPLHLLPAPRP
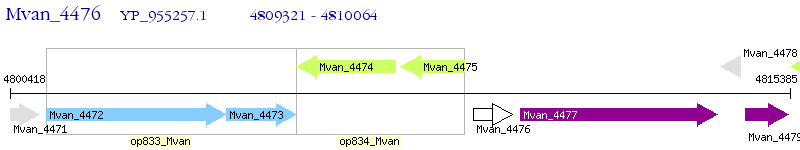
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4476 | - | - | 100% (247) | hypothetical protein Mvan_4476 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1281c | - | 2e-82 | 64.04% (228) | hypothetical protein Mb1281c |
| M. gilvum PYR-GCK | Mflv_2219 | - | 1e-117 | 86.59% (246) | hypothetical protein Mflv_2219 |
| M. tuberculosis H37Rv | Rv1249c | - | 2e-82 | 64.04% (228) | hypothetical protein Rv1249c |
| M. leprae Br4923 | MLBr_01096 | - | 1e-77 | 62.84% (218) | hypothetical protein MLBr_01096 |
| M. abscessus ATCC 19977 | MAB_1394c | - | 3e-70 | 54.82% (228) | hypothetical protein MAB_1394c |
| M. marinum M | MMAR_4193 | - | 1e-83 | 60.08% (243) | hypothetical protein MMAR_4193 |
| M. avium 104 | MAV_1386 | - | 2e-80 | 64.89% (225) | hypothetical protein MAV_1386 |
| M. smegmatis MC2 155 | MSMEG_5048 | - | 1e-105 | 75.90% (249) | hypothetical protein MSMEG_5048 |
| M. thermoresistible (build 8) | TH_1000 | - | 3e-78 | 82.56% (172) | POSSIBLE MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4499 | - | 3e-81 | 58.44% (243) | hypothetical protein MUL_4499 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1281c|M.bovis_AF2122/97 MSARRIRSWKRFDNRSANAAEPDPQLAGTGGRPKVSTRALAQVIERSSRI
Rv1249c|M.tuberculosis_H37Rv MSARRIRSWKRFDNRSANAAEPDPQLAGTGGRPKVSTRALAQVIERSSRI
MMAR_4193|M.marinum_M MSARRTR-----------QSEPAAQPTGIMGRPKASTRALAQVIERSSHI
MUL_4499|M.ulcerans_Agy99 MSARRTR-----------QSEPAAQPTGIMGRPKASTRALAQVIERSSHI
MAV_1386|M.avium_104 MRARRNRFPNR------FFSTSSAAARTDSGRPKVSARALAQVIERSSRI
MLBr_01096|M.leprae_Br4923 ---------------------------------------MAQVIERSVWI
Mvan_4476|M.vanbaalenii_PYR-1 -MFARSRGKG--------QPPAVV-----DEDPGAAAKVLSQIIERGARV
Mflv_2219|M.gilvum_PYR-GCK -MSARSRAKN--------QSPAVT-----DEDAGVAARALSQIIERGARV
MSMEG_5048|M.smegmatis_MC2_155 MVFGRSKSKN--------LPATVEGANGLSDDPGTAAKVLSHIIESSTRV
TH_1000|M.thermoresistible__bu --------------------------------------------------
MAB_1394c|M.abscessus_ATCC_199 -MPMSTRITEP-------QRQLALGGKSKESKPGASGKLLSLVLDQSTKV
Mb1281c|M.bovis_AF2122/97 QGPAAQAYVARLRRAHPGASPAKIVAKLEKRFLSVVTASGAAVGTAATLP
Rv1249c|M.tuberculosis_H37Rv QGPAAQAYVARLRRAHPGASPAKIVAKLEKRFLSVVTASGAAVGAAATLP
MMAR_4193|M.marinum_M QSPAAEAYVARLRRAHPAASPAEIVSKIEKRFLAVVTASGAAVGAAATLP
MUL_4499|M.ulcerans_Agy99 QSPAAEAYMARLRRAHPAASPAEIVSKIEKRFLAVVTASGAAVGAAATLP
MAV_1386|M.avium_104 QGPAAEAYVTRLRNAHPDAGPAEILAKLQKRYLAIVTAGGVAVGAAATIP
MLBr_01096|M.leprae_Br4923 QGPAAEAYVARLRRTHPSASPTEIVAKLEKHYLAALTASGAVVGSVATLP
Mvan_4476|M.vanbaalenii_PYR-1 QGPAVKAYVERLRDNNPGATPAEIIDKLDKKYLAAVMASGAAVGSTAAYP
Mflv_2219|M.gilvum_PYR-GCK QGPAVRAYVARLRENSPEASPADIITKLDTQYMAAAMASGAAVGSAAAYP
MSMEG_5048|M.smegmatis_MC2_155 QAPAVRAYVERLRSQQPDATPAQIVSRLEKHYLAAVMASGAAVGSAAAFP
TH_1000|M.thermoresistible__bu -----------------------------------VVASGAAVGSTAAVP
MAB_1394c|M.abscessus_ATCC_199 QEPAVRAYVQKLRATHPTATPADVVAMLEKRYLATVMGSGAAVGAAAAFP
..*..**:.*: *
Mb1281c|M.bovis_AF2122/97 GIGTLAAWFAAAGEVVVFLEATALFVLALASVHAIPLDHRERRRALVLAV
Rv1249c|M.tuberculosis_H37Rv GIGTLAAWFAAAGEVVVFLEATALFVLALASVHAIPLDHRERRRALVLAV
MMAR_4193|M.marinum_M GIGTLAALSAAAGETAVFLEATALLVLALASVYDIPLDHRERRRALVLAV
MUL_4499|M.ulcerans_Agy99 GIGTLAALSAAAGETAVFLEATALLVLALASVYDHPLDHRERRGALVLAV
MAV_1386|M.avium_104 GIGTLSALSAAAAETVTFLEATAFFVLAAAVVYDIPADHRERRRALVLAV
MLBr_01096|M.leprae_Br4923 GIGTLAAVSANAGETVFFLEATAVFVLTIASVYGIPANHRERRRALVLAV
Mvan_4476|M.vanbaalenii_PYR-1 GIGTLVAMSAVAGETVVFLEATALYVLAVAEVHGIPAEHRERRRALVLSV
Mflv_2219|M.gilvum_PYR-GCK GIGTLVAMSAVAGETVVFLEATTLYVLAMAEVHGIPAEHRERRRALVLSV
MSMEG_5048|M.smegmatis_MC2_155 GIGTLAALSAVAGETVVFLEATSVFVLAVAEVHGIPADHRERRRALVLSV
TH_1000|M.thermoresistible__bu GVGTLAALSAVTGETAVFLEATAVFVLAVAEVHGIPLEERERRRALVLAV
MAB_1394c|M.abscessus_ATCC_199 GVGTLAALSATAGEAVLFLEATSLFTLALAAVHDIELAKIEHRHTLVLAV
*:*** * * :.*.. *****:. .*: * *: . *:* :***:*
Mb1281c|M.bovis_AF2122/97 LVGDN-TTAVADLLGPGRTSG-GWVSETMASLPLPAISSLNSRMLKYVVK
Rv1249c|M.tuberculosis_H37Rv LVGDN-TTAVADLLGPGRTSG-GWVSETMASLPLPAISSLNSRMLKYVVK
MMAR_4193|M.marinum_M LVGDNSKNAVAEVIGSGRTSG-VWVSESMASLPLPAISKLNTRMFKYFVK
MUL_4499|M.ulcerans_Agy99 LVGDNSKNAVAEVIGSGRTSG-VWVSESMASLPLPAISKLNTRMFKYFVK
MAV_1386|M.avium_104 LVGDNSERAVAQLIGPGRTQG-GWVSESLAAVPLSSMSRLNSRLLKSWVR
MLBr_01096|M.leprae_Br4923 LAGDDTRLTIGELIGPGRTNG-GWLLEGMASLPLSTWSQLHTRMLRYAAK
Mvan_4476|M.vanbaalenii_PYR-1 LVGEDSKRAVADLLGSGRTSG-AWVSDGAATLPLPALSQLNSRLLRYFVK
Mflv_2219|M.gilvum_PYR-GCK LVGEDSKRAVADLLGPGRTSG-AWVSDGAATLPLPALSQLNTRLLRYFVK
MSMEG_5048|M.smegmatis_MC2_155 LVGEDSKRALADLLGTGRTSG-AWLSDGAATLPLPAVSQLNSRLVRYFVK
TH_1000|M.thermoresistible__bu LVGDEGKGAVRDLLGRGRTSG-AWISEGAATLPLPTVSQLNSRLLRYFVK
MAB_1394c|M.abscessus_ATCC_199 LLGEDGETTVSDVLGKERTVGGGWLAEATSTLPVPALSHLNATLLKRFTK
* *:: :: :::* ** * *: : :::*:.: * *:: :.: .:
Mb1281c|M.bovis_AF2122/97 RFALKRGALMFGKLVPMGIGAIIGAIGNRLVGKKLVRNARSAFGTPPARW
Rv1249c|M.tuberculosis_H37Rv RFALKRGALMFGKLVPMGIGAIIGAIGNRLVGKKLVRNARSAFGTPPARW
MMAR_4193|M.marinum_M RFALKRGVLLFGKLLPAGIGAIIGAIGNRLVGRKLVRNARSAFGAPPARW
MUL_4499|M.ulcerans_Agy99 RFALKRGVLLFGKLLPAGIGAIIGAIGNRLVGRKLVRNARSAFGAPPTRW
MAV_1386|M.avium_104 RYTLRRGALLFGKLLPVGIGVVVGAVGNYLAGKKIIRNANRAFGAPPARW
MLBr_01096|M.leprae_Br4923 RCTVRRGALMFGKILPIGIGAAVGGAGNRVVGKKIISNTRNAFGTAPSRW
Mvan_4476|M.vanbaalenii_PYR-1 RYTLKRGALAFGKMLPVGIGAVVGGVGNRFMGKKIIANAHNAFGAAPARW
Mflv_2219|M.gilvum_PYR-GCK RYTLKRGALAFGKMLPVGIGAVVGGVGNRLMAKKIVGNAHTAFGNPPARW
MSMEG_5048|M.smegmatis_MC2_155 RYTLKRGAMAFGKLLPVGIGALIGGVGNRMMGKKIVANAAKAFGAPPPRW
TH_1000|M.thermoresistible__bu RYTLKRGALAFGKALPVGVGAVVGGVGNRMMGKKIIGNARTAFGAAPPRW
MAB_1394c|M.abscessus_ATCC_199 KFAVKRGAMAAGKLLPAGIGAVIGAGGNRFIGKRIITNARNAFGEPPTAW
: :::**.: ** :* *:*. :*. ** . .:::: *: *** .*. *
Mb1281c|M.bovis_AF2122/97 PVTLHVLPTVRDAS----
Rv1249c|M.tuberculosis_H37Rv PVTLHVLPTVRDAS----
MMAR_4193|M.marinum_M PVTLHVLPNVRDAS----
MUL_4499|M.ulcerans_Agy99 PVTLHVLPNVRDAS----
MAV_1386|M.avium_104 PRALHLVPPIHEAG----
MLBr_01096|M.leprae_Br4923 PATLILLPTVHNAG----
Mvan_4476|M.vanbaalenii_PYR-1 PSPLHLLPAPRP------
Mflv_2219|M.gilvum_PYR-GCK PSPLHLLPAPRD------
MSMEG_5048|M.smegmatis_MC2_155 PATLHVLPSAQT------
TH_1000|M.thermoresistible__bu PAPLHLLPPLPESGTDST
MAB_1394c|M.abscessus_ATCC_199 PVQLRLVAPDS-------
* * ::.