For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPPTLQSFPPLAAPGARILILGNMPGVASLQAQRYYAHTRNAFWGITGELFGFDPAAPYDHRVAVLTAA GVAVWDVLRHCRRAGSLDSAVEPDSMVPNDFASFFADHPAITHVYFNGAAAEKNYGRLVTAPATVSYTRL PSTSPAHTMSFDRKLAAWRQITAAVRA
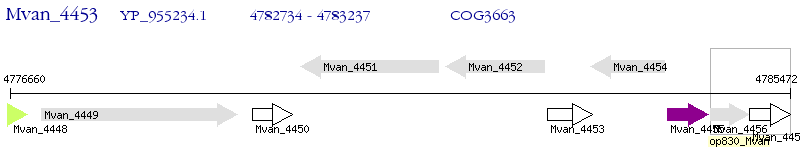
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4453 | - | - | 100% (167) | hypothetical protein Mvan_4453 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2242 | - | 4e-64 | 77.18% (149) | hypothetical protein Mflv_2242 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1413 | - | 2e-51 | 60.76% (158) | T/U mismatch-specific DNA glycosylase |
| M. smegmatis MC2 155 | MSMEG_5024 | - | 1e-50 | 61.04% (154) | T/U mismatch-specific DNA glycosylase |
| M. thermoresistible (build 8) | TH_1843 | - | 6e-45 | 62.07% (145) | T/U mismatch-specific DNA glycosylase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4453|M.vanbaalenii_PYR-1 MTPPTLQSFPPLAAPGARILILGNMPGVASLQAQRYYAHTRNAFWGITGE
Mflv_2242|M.gilvum_PYR-GCK MG----------ANP--RILILGNMPGVASLRAQRYYWYDRNAFWRITAA
MSMEG_5024|M.smegmatis_MC2_155 MSS-VLHGLPPIVGEAPRVLILGNMPSVMSLASSEYYGNPRNAFWRIMGS
TH_1843|M.thermoresistible__bu ------------------------VPGVQSLRAQRYYETPRNAFWRIIGE
MAV_1413|M.avium_104 MSTPLLRGFPPVVDERARTLILGSFPSAQSLLTGQYYANPRNAFWSITGE
.*.. ** : .** ***** * .
Mvan_4453|M.vanbaalenii_PYR-1 LFGFDPAAPYDHRVAVLTAAGVAVWDVLRHCRRAGSLDSAVEPDSMVPND
Mflv_2242|M.gilvum_PYR-GCK LFDFDPASAYDERVAALTGAGVAVWDVLKHCRRVGSLDSAVEPDSMVPND
MSMEG_5024|M.smegmatis_MC2_155 LYGFAPDAAYSERVSAVTSHGIAVWDVLRSCRRVGSLDSAVERDSMVPND
TH_1843|M.thermoresistible__bu LLGFEPDAPYDVRLDAVRARSVAVWDVLRCCRRKGSLDSAVERDSMQPND
MAV_1413|M.avium_104 LFGFDAAAPYPRRLAQLRRHRIALWDVLHACRRAGSADSAIEPNSLVVND
* .* . :.* *: : :*:****: *** ** ***:* :*: **
Mvan_4453|M.vanbaalenii_PYR-1 FASFFADHPAITHVYFNGAAAEKNYGRLVTAPA----TVSYTRLPSTSPA
Mflv_2242|M.gilvum_PYR-GCK FVAFFETHRTLTHVYFNGAAAERNYARLVGGPD----TLGYTRLPSTSPA
MSMEG_5024|M.smegmatis_MC2_155 FAGFFAAHPSLERVVFNGAAAEANYRRLVGEVP-----LPYVRMPSTSPA
TH_1843|M.thermoresistible__bu FGAFFAEHQTITRVVFNGAAAEANFIRLVGADPGIESDVEFVRAPSTSPA
MAV_1413|M.avium_104 FGEFFAEHPGITRVYFNGAKAAELYRRLATAPD----HVCFQRLPSTSPA
* ** * : :* **** * : **. : : * ******
Mvan_4453|M.vanbaalenii_PYR-1 HTMSFDRKLAAWRQITAAVRA-
Mflv_2242|M.gilvum_PYR-GCK QTMSFDAKLDAWRQITAPART-
MSMEG_5024|M.smegmatis_MC2_155 QTMRFEDKLSAWRVELER----
TH_1843|M.thermoresistible__bu QTMRYDDKLRAWRSALTPVCLS
MAV_1413|M.avium_104 HVMAPGAKLAAWAVLRNSA---
:.* ** **