For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLPGSVAEIHASPEGPEIGAFFDLDGTLVAGFTGVVMTQDRLRRRQMSVGEFIGMVQAGLNHQLGRSEFE DLIGKGARMLRGNSVDDIDELAERLFVQKIVGRIYPEMREIVRAHMARGHTVVLSSSALTVQVEPVARFL GINNVLSNKFETDDDGLITGEVQRPIIWGPGKARAVQEFAAANDIDLSKSYFYADGDEDVALMYLVGNPR PTNPAGKMAAVAAKRGWPILRFSSRSGASPASQVRTAVGIATMVPIAAGAIGVGLLTRNKRTGVNFFTSM FGRTLLNTVGINLQVLGKENLTAQRPAVFIFNHRNQADPLIAGRLVNDNFTSVGKKELENDPIVGTMGKI MDAAFIDRDDPQKAVEGLHKVEELARKGLSILIAPEGTRLDTTEVGPFKKGPFRIAMSVGIPIVPIVIRN AEVIAARDSSTFNPGTVDVVVYPPIPVDDWTRENLSERIDEVRQLYIDTLKDWPHDELPTPELYRRAKPA KKSTARKTPAKKAPAKKAAAKAPTKRAGGTKGRP*
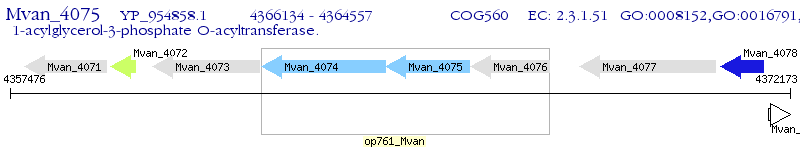
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4075 | - | - | 100% (525) | HAD family hydrolase |
| M. vanbaalenii PYR-1 | Mvan_3004 | - | 1e-74 | 55.42% (240) | HAD family hydrolase |
| M. vanbaalenii PYR-1 | Mvan_0843 | - | 5e-21 | 27.92% (265) | HAD family hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2508c | plsC | 0.0 | 69.94% (529) | bifunctionnal putative L-3-phosphoserine |
| M. gilvum PYR-GCK | Mflv_2564 | - | 0.0 | 87.93% (522) | HAD family hydrolase |
| M. tuberculosis H37Rv | Rv2483c | plsC | 0.0 | 69.75% (529) | bifunctionnal putative L-3-phosphoserine |
| M. leprae Br4923 | MLBr_01245 | - | 0.0 | 66.16% (526) | putative transferase |
| M. abscessus ATCC 19977 | MAB_2455c | - | 7e-74 | 52.61% (249) | hypothetical protein MAB_2455c |
| M. marinum M | MMAR_3834 | plsC | 0.0 | 70.17% (533) | bifunctional transmembrane phospholipid biosynthesis enzyme |
| M. avium 104 | MAV_1687 | - | 0.0 | 68.93% (544) | acyltransferase family protein |
| M. smegmatis MC2 155 | MSMEG_4704 | - | 0.0 | 82.84% (507) | acyltransferase family protein |
| M. thermoresistible (build 8) | TH_1038 | - | 0.0 | 77.13% (529) | POSSIBLE TRANSMEMBRANE PHOSPHOLIPID BIOSYNTHESIS BIFUNCTIONNAL |
| M. ulcerans Agy99 | MUL_3764 | plsC | 0.0 | 70.62% (531) | bifunctional transmembrane phospholipid biosynthesis enzyme |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2508c|M.bovis_AF2122/97 MSAADEQGEERATRKSAPDLRLPGSVAEILASPAGPKVGAFFDLDGTLVA
Rv2483c|M.tuberculosis_H37Rv MSAADEQGEERATRKSAPDLRLPGSVAEILASPAGPKVGAFFDLDGTLVA
MMAR_3834|M.marinum_M MSAAEEPGDGREQDKAAGDLRLPGSVAEIMASPAGPKVGAFFDLDGTLVA
MUL_3764|M.ulcerans_Agy99 MSAAEEPGDGREQDKAAGDLRLPGSVAEIMASPAGPKVGAFFDLDGTLVA
MAV_1687|M.avium_104 -------------------MRLPGSVAEIMASPPGPRIGAFFDLDGTLVA
MLBr_01245|M.leprae_Br4923 MSAFDENG----TPKSGPEVRLPGSVAEILASPAGPQVGAFFDLDGTLVA
Mvan_4075|M.vanbaalenii_PYR-1 -------------------MRLPGSVAEIHASPEGPEIGAFFDLDGTLVA
Mflv_2564|M.gilvum_PYR-GCK ---MTSEIP------VNRQMRLPGSVAEIQASPAGAQIGAFFDLDGTLVA
MSMEG_4704|M.smegmatis_MC2_155 -------------------MRLPGTLAEIEASPEGPEIGAFFDLDGTLVA
TH_1038|M.thermoresistible__bu ---MTAEDRRAERDEEERTLRLPGSVAEIEASPPGPEIGAFFDLDGTLVA
MAB_2455c|M.abscessus_ATCC_199 ----------------MTANSTPHPVADIALSPVGSEVGAFFDLDGTLVA
* .:*:* ** *..:************
Mb2508c|M.bovis_AF2122/97 GFTAVILTQERLRRRDMGVGELLGMVQAGLNHTLGRIEFEDLIGKAAAAL
Rv2483c|M.tuberculosis_H37Rv GFTAVILTQERLRRRDMGVGELLGMVQAGLNHTLGRIEFEDLIGKAAAAL
MMAR_3834|M.marinum_M GFTAVILTQERLRRRDIGVGELLSMVQAGLNHTLGRIEFEQLINTASSAL
MUL_3764|M.ulcerans_Agy99 GFTAVILTQERLRRRDIGVGELLSMVQAGLNHTLGRIEFEQLINTASSAL
MAV_1687|M.avium_104 GFTAVILTQERLLRRDMGVGELLGMVQAGLSHTLGRIEFEDLIGKAAAAL
MLBr_01245|M.leprae_Br4923 GFTAVILTHERLRRCDMGVGELLSMIQAGLNHTLGRIEFEDLIGKVSSAL
Mvan_4075|M.vanbaalenii_PYR-1 GFTGVVMTQDRLRRRQMSVGEFIGMVQAGLNHQLGRSEFEDLIGKGARML
Mflv_2564|M.gilvum_PYR-GCK GFTGVVMTQDRLRRRQMSVGEFIGMVQAGLNHQLGRSEFEDLIGKGARML
MSMEG_4704|M.smegmatis_MC2_155 GFTGVLMTQDRLRRRQMSVGEFIGMVQAGLNHRLGRSEFEDLIGKGARML
TH_1038|M.thermoresistible__bu GFTGVIMTRERLRRRELSVGEFIGLVQAGLNHQLGRAEFEDLIGRGARML
MAB_2455c|M.abscessus_ATCC_199 GFTPTAHARDRMRRRQASVGEVLGVLEATFRYKLGRMEFERLVVRAAGYL
*** . :::*: * : .***.:.:::* : : *** *** *: : *
Mb2508c|M.bovis_AF2122/97 AGRLLTDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
Rv2483c|M.tuberculosis_H37Rv AGRLLTDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
MMAR_3834|M.marinum_M RGRQLVDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
MUL_3764|M.ulcerans_Agy99 RGRQLVDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
MAV_1687|M.avium_104 AGRLIDDLEEIGERMFVQRIESRIYPEMRELVRAHMARGHTVVLSSSALT
MLBr_01245|M.leprae_Br4923 RGRLLTDLEEIGERLFAQRIESRIYPEMRKLVQAHVARGHTVVLSSSALT
Mvan_4075|M.vanbaalenii_PYR-1 RGNSVDDIDELAERLFVQKIVGRIYPEMREIVRAHMARGHTVVLSSSALT
Mflv_2564|M.gilvum_PYR-GCK RGNSLDDVDELAERLFVQKIVGRIYPEMREIVRAHMNRGHTVVLSSSALT
MSMEG_4704|M.smegmatis_MC2_155 RGNSLSDLDELGERLFVQYVQGRIYPEMRAMVRAHMARGHTVVLSSSALT
TH_1038|M.thermoresistible__bu RGSSLSDIDELAERLFVQKIQNRIYPEMRELVRAHQARGHTVVLSSSALT
MAB_2455c|M.abscessus_ATCC_199 RGDSLAELEAVGERIFHQHNAAMIYPQMRERVSAHQNQGHTVVLSSSALT
* : ::: :.**:* * ***:** * ** :************
Mb2508c|M.bovis_AF2122/97 IQVGPVARFLGINNMLTNKFETNEDGILTGGVLKPILWGPGKATAVQRFA
Rv2483c|M.tuberculosis_H37Rv IQVGPVARFLGINNMLTNKFETNEDGILTGGVLKPILWCPGKATAVQRFA
MMAR_3834|M.marinum_M IQVNPVARFLGIVNMLTNKFETNEDGLLTGGVVKPILWGPGKAAAVQRFA
MUL_3764|M.ulcerans_Agy99 IQVNPVARFLGIVNMLTNKFETNEDGLLTGGVVKPILWGPGKAAAVQRFA
MAV_1687|M.avium_104 IQVNPVARFLGIPNTLTNKFETTEDGILTGGLQKPILWGPGKAAAVQRFA
MLBr_01245|M.leprae_Br4923 IQVGPVARFLGIAHMLTNKFEINEDGMLTGGVVKPILWGPGKAAAVQRFA
Mvan_4075|M.vanbaalenii_PYR-1 VQVEPVARFLGINNVLSNKFETDDDGLITGEVQRPIIWGPGKARAVQEFA
Mflv_2564|M.gilvum_PYR-GCK VQVEPVARFLGIDNVLSNKFEIDDDGLITGEVQRPIIWGPGKARAVQEFA
MSMEG_4704|M.smegmatis_MC2_155 VQVEPVARFLGIDNVLSNKFETDEDGLITGEVRTPIIWGPGKARAVQEFA
TH_1038|M.thermoresistible__bu VQVEPVARYLGIENILSNKFEVDENGCITGEVNRPIIWGPGKARAVQAFA
MAB_2455c|M.abscessus_ATCC_199 IHAEPVARHLGITHVLCNHFDVDNRGLLTGDIRKPIVWGRNKASVVRSFC
::. ****.*** : * *:*: : * :** : **:* .** .*: *.
Mb2508c|M.bovis_AF2122/97 AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
Rv2483c|M.tuberculosis_H37Rv AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
MMAR_3834|M.marinum_M AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
MUL_3764|M.ulcerans_Agy99 AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
MAV_1687|M.avium_104 AEHDIDLKESYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
MLBr_01245|M.leprae_Br4923 AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
Mvan_4075|M.vanbaalenii_PYR-1 AANDIDLSKSYFYADGDEDVALMYLVGNPRPTNPAGKMAAVAAKRGWPIL
Mflv_2564|M.gilvum_PYR-GCK SVNGIDLSRSYFYADGDEDVALMYLVGNPRPTNPAGKMAAVAAKRGWPIL
MSMEG_4704|M.smegmatis_MC2_155 AANGVDLSKSYFYADGDEDVALMYLVGNPRPTNPEGKLAAVAAKRGWPIL
TH_1038|M.thermoresistible__bu AERGIDLSRSYFYADGDEDVALMYLVGNPRPTNPAGKMASVAAKRGWPIL
MAB_2455c|M.abscessus_ATCC_199 ETNGVDLQRSYFYADGEEDIDLMALVGEPRPVNPRGRLAVMAAEQGWPVL
..:**. *******:**: ** ***:***.** *::* :* .:***:*
Mb2508c|M.bovis_AF2122/97 KFNSRGGVGIRRQLRTLAGLSTIVPVAAGAVGIGVLTGSRRRGVNFFTST
Rv2483c|M.tuberculosis_H37Rv KFNSRGGVGIRRQLRTLAGLSTIVPVAAGAVGIGVLTGSRRRGVNFFTST
MMAR_3834|M.marinum_M KFNSRGGVGLRRQIRTLAGFSTMFPVAAGAVGIGLLTGNRRRGVNFFTSN
MUL_3764|M.ulcerans_Agy99 KFNSRGGVGLRRQIRTLAGFSTMFPVAAGAVGIGLLTGNRRRGVNFFTSN
MAV_1687|M.avium_104 RFSSRGPVGLRRQLRTLAGFGSMFPVAAGAVGVGVLTRNRRRGVNFFTST
MLBr_01245|M.leprae_Br4923 KFNSRGGVGIGAQLRTLVGLSSVLPLVAGAVGIGVLTGSRRRGANFFTSV
Mvan_4075|M.vanbaalenii_PYR-1 RFSSRSGASPASQVRTAVGIATMVPIAAGAIGVGLLTRNKRTGVNFFTSM
Mflv_2564|M.gilvum_PYR-GCK KFSSRSGASPASQVRTAVGIASMVPIAAGALGLGLLTRNKRTGVNFFTSM
MSMEG_4704|M.smegmatis_MC2_155 RFTSRSGSSPMAQLRTVAGVASLMPVAGAAIGWGLLNRNRRRGVNFFTST
TH_1038|M.thermoresistible__bu RFTSRRGSNPVSQVRTAVGVASLLPVAAGAVGVGLLSRSRRAGVNFLTSV
MAB_2455c|M.abscessus_ATCC_199 RLEDPRPRGMRPSLRRLAGLARP---------------------------
:: . . .:* .*..
Mb2508c|M.bovis_AF2122/97 FSQLLLATSGVHLNVIGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
Rv2483c|M.tuberculosis_H37Rv FSQLLLATSGVHLNVIGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
MMAR_3834|M.marinum_M FSQLLLATSGVHLNVVGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
MUL_3764|M.ulcerans_Agy99 FSQLLLATSGVHLNVVGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
MAV_1687|M.avium_104 FSQLSLAIAGVHLNVIGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
MLBr_01245|M.leprae_Br4923 FPPLVLAAAGVHLNVIGNENLTMQRPAVFIYNHRNQIDPLIAGALVRGNW
Mvan_4075|M.vanbaalenii_PYR-1 FGRTLLNTVGINLQVLGKENLTAQRPAVFIFNHRNQADPLIAGRLVNDNF
Mflv_2564|M.gilvum_PYR-GCK FGRTLLDTIGINLQVLGRENLTAQRPAVFIFNHRNQADPLIAGRLINEDF
MSMEG_4704|M.smegmatis_MC2_155 WSRVLLATTGVNLNVLGEENLTAQRPAVFIFNHRNQVDPIIAGTLVRDNF
TH_1038|M.thermoresistible__bu WGKLLLAATGVELNVLGEENLTAERPAVFIFNHRNQVDPVIAGRLVKDNF
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 VGVGKKELASDPIMGTLGKLLDGVFIDRDDPVAAVETLHTVEERARNGLS
Rv2483c|M.tuberculosis_H37Rv VGVGKKELASDPIMGTLGKLLDGVFIDRDDPVAAVETLHTVEERARNGLS
MMAR_3834|M.marinum_M VGVGKKELQNDPIMGTLGKVLDGVFIDRDDPASAVETLHTVEERAKNGLS
MUL_3764|M.ulcerans_Agy99 VGVGKKELQNDPIMGTLGKVLDGVFIDRDDPASAVETLHTVEERAKNGLS
MAV_1687|M.avium_104 VAVGKKELEKDPVMGTLGKLLDGVFIDRDDPAAAVETLHTVEDRARNGLS
MLBr_01245|M.leprae_Br4923 IAVAKKELAKNPIIRALGKLTDGVFIDRDDPVGAVETLHAVEDRARKGLS
Mvan_4075|M.vanbaalenii_PYR-1 TSVGKKELENDPIVGTMGKIMDAAFIDRDDPQKAVEGLHKVEELARKGLS
Mflv_2564|M.gilvum_PYR-GCK TTVGKKELENDPIVGTIGKILDAAFIDRDDPKKAVEGLKKVEELARKGLS
MSMEG_4704|M.smegmatis_MC2_155 TGVGKKELANDPLVGTLGRVMDAAFIDRDDPVKAVEGLHKVEELAREGIS
TH_1038|M.thermoresistible__bu TMVGKKELADDPLLGMLGKVMDVAFIDRSDPTAAVEGLRKVEELAAKGVS
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 IVIAPEGTRLDTTEVGSFKKGPFRIAMAAKIPIVPIVIRNAEIVASRNST
Rv2483c|M.tuberculosis_H37Rv IVIAPEGTRLDTTEVGSFKKGPFRIAMAAKIPIVPIVIRNAEIVASRNST
MMAR_3834|M.marinum_M IVIAPEGTRLDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAEIVASRNST
MUL_3764|M.ulcerans_Agy99 IVIAPEGTRLDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAEIVASRNST
MAV_1687|M.avium_104 ILIAPEGTRVDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAELVAARNST
MLBr_01245|M.leprae_Br4923 ILMAPEGTRLDTTEVGPFKKGPFRIAMAVGIPIVPIVFRNAEIVAARNSN
Mvan_4075|M.vanbaalenii_PYR-1 ILIAPEGTRLDTTEVGPFKKGPFRIAMSVGIPIVPIVIRNAEVIAARDSS
Mflv_2564|M.gilvum_PYR-GCK ILIAPEGTRLDTTEVGEFKKGPFRIAMSVNIPIVPIVIRNAEVIAARDSS
MSMEG_4704|M.smegmatis_MC2_155 ILIAPEGTRLDTTAVGPFKKGAFRIAMAAGIPVVPIVIRNAEVIAARDSS
TH_1038|M.thermoresistible__bu ILIAPEGTRVDTNEVGPFKKGAFRIAMATGIPIVPIVIRNAEVVAARNST
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 TINPGTVDVAVFPPIPVDDWTLDALPDRIAEVRQLYLDTLADWPVDGLPA
Rv2483c|M.tuberculosis_H37Rv TINPGTVDVAVFPPIPVDDWTLDALPDRIAEVRQLYLDTLADWPVDGLPA
MMAR_3834|M.marinum_M TINPGTVDIAVFPPIPVDDWTLDTLPDRIAEVRQLYLDTLKDWPVDELPE
MUL_3764|M.ulcerans_Agy99 TINPGTVDIAVFPPIPVDDWTLDTLPDRIAEVRQLYLDTLKDWPVDELPE
MAV_1687|M.avium_104 VINPGTVDVAVFPPISVQDWTLDTLPERIAEVRQLYLDTLANWPVGELPE
MLBr_01245|M.leprae_Br4923 TINPGMVDVAVLPAIPVDDWTLDALSERIAEVRQLYLDILADWPVDELPK
Mvan_4075|M.vanbaalenii_PYR-1 TFNPGTVDVVVYPPIPVDDWTRENLSERIDEVRQLYIDTLKDWPHDELPT
Mflv_2564|M.gilvum_PYR-GCK TFNPGLVDVVVYPPIPVDDWTQEDLPSRIEEIRQLYLDTLADWPHDELKL
MSMEG_4704|M.smegmatis_MC2_155 TFNPGTVDVAVYPPIPVDDWTVENLSERIEQVRQIYLDTLASWPHDRLPE
TH_1038|M.thermoresistible__bu TFNPGKVDVVVYPPIPVDDWTVEELPERIAEVRQLYLDTLKAWPRKHLPL
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 VDLYAEQKAARKARAQVAKATAKRV-------PAKKAPAKSAAN------
Rv2483c|M.tuberculosis_H37Rv VDLYAEQKAARKARAQVAKATAKRV-------PAKKAPAKSAAN------
MMAR_3834|M.marinum_M VNLYAEEKAAKKAKAQLAKASAKETPAK--KAPAKKAPAKKAAAKKETSA
MUL_3764|M.ulcerans_Agy99 VNLYAEEKAAKKAKAQLAKASAKET-------PAKKAPAKKAAAKKET--
MAV_1687|M.avium_104 VDLYAEKKAAQKARSRAAKSPATSPAKTPAKSPAKRPAKKSAAKKT----
MLBr_01245|M.leprae_Br4923 AALYTRATKATVKKAEPKKGGRTSAAKT----TAKKTATKATAAKT----
Mvan_4075|M.vanbaalenii_PYR-1 PELYRRAK-PAKKSTARKTPAKKAP--------AKKAAAKAPTK------
Mflv_2564|M.gilvum_PYR-GCK PDLYTRSQ-PAKKRTPRKAASKKIPTKT-ATKTAKKAQPKTTKK------
MSMEG_4704|M.smegmatis_MC2_155 FDIYSPRK-AAKKT-------------------ARKAAKKAPAK------
TH_1038|M.thermoresistible__bu PELYTRPRGPAKKAPVKKSQAKKTQ--------AKKTPVKKTQA------
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 ----------KGAAATKAATKKASPKAKPSESKIAGKD------------
Rv2483c|M.tuberculosis_H37Rv ----------KGAAATKAATKKASPKAKPSESKIAGKD------------
MMAR_3834|M.marinum_M KKAPAKKAPAKKAAAKKAPAKKATAKKAPAKKAPAKKVPESAAAKAATPA
MUL_3764|M.ulcerans_Agy99 ---PAKKAPAKKAPAKKAPAKKATAKKAPANKAPAKKVPESAAAKAATRA
MAV_1687|M.avium_104 ----------AATAAAKTRAKKTAAKPNPAAPNPEAAASHN---------
MLBr_01245|M.leprae_Br4923 ----------ANTAVTKSMLTPPTRISDKENPKVPKVVS-----------
Mvan_4075|M.vanbaalenii_PYR-1 -----------RAGGTKG---------RP---------------------
Mflv_2564|M.gilvum_PYR-GCK -----------TQPKTKATRASSKVSDRPDADPPEPAEK-----------
MSMEG_4704|M.smegmatis_MC2_155 -----------GRP------------------------------------
TH_1038|M.thermoresistible__bu -----------KKAQAKKAQAGKSADHPPGLGTQERS-------------
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 -----------GEAS---------------------------ASPSSSAK
Rv2483c|M.tuberculosis_H37Rv -----------GEAS---------------------------ASPSSSAK
MMAR_3834|M.marinum_M TDTELPAPKPVGEASNPTEIAPEPIAHDGDAQQLGAGRSDAAESSSSRPK
MUL_3764|M.ulcerans_Agy99 TDTELSAPKPVGEASNPTEIAPEPIAHDGDAQQLGAGRSDAAESSSSRPK
MAV_1687|M.avium_104 -----------GEVSPSRATDPIG-----------------ADPPEAPPR
MLBr_01245|M.leprae_Br4923 -----------------------------------------QDLVVKQPR
Mvan_4075|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2564|M.gilvum_PYR-GCK --------------------------------------------PADSRE
MSMEG_4704|M.smegmatis_MC2_155 --------------------------------------------------
TH_1038|M.thermoresistible__bu --------------------------------------------------
MAB_2455c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2508c|M.bovis_AF2122/97 GRS
Rv2483c|M.tuberculosis_H37Rv GRS
MMAR_3834|M.marinum_M GRP
MUL_3764|M.ulcerans_Agy99 GRP
MAV_1687|M.avium_104 GRP
MLBr_01245|M.leprae_Br4923 ERS
Mvan_4075|M.vanbaalenii_PYR-1 ---
Mflv_2564|M.gilvum_PYR-GCK GTS
MSMEG_4704|M.smegmatis_MC2_155 ---
TH_1038|M.thermoresistible__bu ---
MAB_2455c|M.abscessus_ATCC_199 ---