For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSFQLAVCAEMVFTDLPILDRVRRIDELGFAAEIWSWHDKDLAALATTGATFTSMTGYLHGDLIDPATCD EVVRTAERSIRAAETLGVTRLNLHTAELVDGRAARPRQRSTGQMWLTAARTLERVGELGAASGVTFCVEN LNTIVDHPGVPLARAKDTLALVEGVGHPNVKMMLDLYHAQIGEGNLIELIHRCKDAIGEIQVADVPGRCE PGTGEINYQAIANALRDIGFDGTIGMEAWASNAGVAGSEAALAAFRTAFA
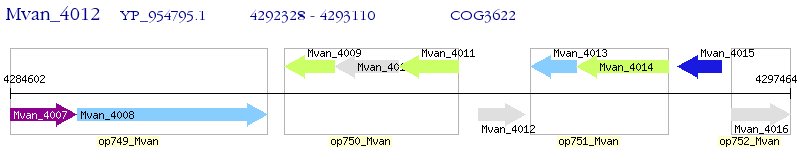
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4012 | - | - | 100% (260) | xylose isomerase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2604 | - | 1e-125 | 83.01% (259) | xylose isomerase domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4653 | - | 1e-106 | 74.51% (255) | AP endonuclease, family protein 2 superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4012|M.vanbaalenii_PYR-1 MSFQLAVCAEMVFTDLPILDRVRRIDELGFAAEIWSWHDKDLAALATTGA
Mflv_2604|M.gilvum_PYR-GCK MTFQLAVCAEMVFQDLPLLDRVERIHELGFAVEIWSWHDKDLAALAATGA
MSMEG_4653|M.smegmatis_MC2_155 MSFDLAVCAEMVFTDLDIVERVRRISDLGFAVEIWSFADKDTDALAATGA
*:*:********* ** :::**.** :****.****: *** ***:***
Mvan_4012|M.vanbaalenii_PYR-1 TFTSMTGYLHGDLIDPATCDEVVRTAERSIRAAETLGVTRLNLHTAELVD
Mflv_2604|M.gilvum_PYR-GCK TFTSMTGYLHGDLIDPQSCDEVVRTAELSVTAAEALGVTRLNLHTAELVD
MSMEG_4653|M.smegmatis_MC2_155 KFSSMTGYLHGDLYDPDGAREVVRTAQEAVKAAEVLGVPRLVVHPGELVN
.*:********** ** . ******: :: ***.***.** :*..***:
Mvan_4012|M.vanbaalenii_PYR-1 GRAARPRQRSTGQMWLTAARTLERVGELGAASGVTFCVENLNTIVDHPGV
Mflv_2604|M.gilvum_PYR-GCK GRAARPRYRADGRMWATALRTLERIGELGARAGVTFCVENLNTVVDHPGV
MSMEG_4653|M.smegmatis_MC2_155 GQAARPRQRVTGDMWLAALRGLEALGELGADAGVTFCLENLNTIVDHPGV
*:***** * * ** :* * ** :***** :*****:*****:******
Mvan_4012|M.vanbaalenii_PYR-1 PLARAKDTLALVEGVGHPNVKMMLDLYHAQIGEGNLIELIHRCKDAIGEI
Mflv_2604|M.gilvum_PYR-GCK PLARAKDTLALVEGVGHAHVKMMLDLYHAQIGEGNLCELVRRCGDAIGEI
MSMEG_4653|M.smegmatis_MC2_155 PLARAKDTLALIEGVGHPNVKMMLDLYHAQIGEGNLVELVRRAGPAIGEI
***********:*****.:***************** **::*. *****
Mvan_4012|M.vanbaalenii_PYR-1 QVADVPGRCEPGTGEINYQAIANALRDIGFDGTIGMEAWASNAGVAGSEA
Mflv_2604|M.gilvum_PYR-GCK QIADVPGRCEPGTGEINYPAIAATLRDIGYTGTVGMEGWASQPGTPGSEA
MSMEG_4653|M.smegmatis_MC2_155 QVADVPGRCEPGTGEIHYPAIAKALRDSGYRGTVGMEAYASG----DSVA
*:**************:* *** :*** *: **:***.:** .* *
Mvan_4012|M.vanbaalenii_PYR-1 ALAAFRTAFA-----------
Mflv_2604|M.gilvum_PYR-GCK ALSAFRAAFEPDYRSTSVVSK
MSMEG_4653|M.smegmatis_MC2_155 ALEAFRAAFTLP---------
** ***:**