For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRSITIDERRARLARRHHLAPGPDGAPSVHAVTGRLVGLHATDPATPHLSLWARLPGYTVADLNAALYER RSVVKQLAMRRTLWVIRAEDLPAVQSAAGDRVATNETRRLAADAQNAGVARDGHAWLEAACAAVVRHLRG AGPCTARELREALPELTGTYDPAPGKPYGGEGHLAPRVLTVLSARGLIVRGPNDGGWTTSRPRWAAAGSW LGPADPVSTDQARAELVRRWLHAFGPATVDDLKWWFGATLGWARQALSDVDAVEVAVEGSAGTSGFVLPG DDGPEPDVEPWCALLPGLDVTTMGWAGRDWYLGPHRGAVFDRNGNAGPTVWVDGRVVGAWRQDDDGRVEL MLLEDVGRPALRALTARAEELTAWLGGVQVKPRFPSPASKSAARR
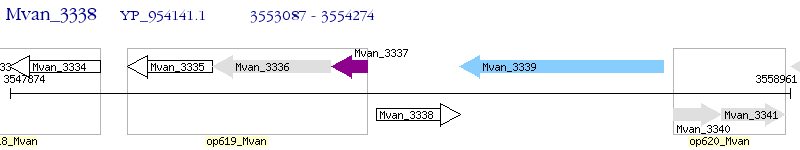
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3338 | - | - | 100% (395) | hypothetical protein Mvan_3338 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3547 | - | 1e-177 | 75.50% (400) | hypothetical protein Mflv_3547 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3806 | - | 1e-147 | 65.98% (391) | hypothetical protein MSMEG_3806 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3338|M.vanbaalenii_PYR-1 -------MRSITIDERRARLARRHHLAPGPDGAPSVHAVTGRLVGLHATD
Mflv_3547|M.gilvum_PYR-GCK MEVRSASMRFFTIDERRARLARRHHLIPGDHRADSVDAVTARLVALHATD
MSMEG_3806|M.smegmatis_MC2_155 -------MRVFTIAERRTRLARRHFLSP-ENPAASPAAAARSFVGLHATD
** :** ***:******.* * . * * *.: :*.*****
Mvan_3338|M.vanbaalenii_PYR-1 PATPHLSLWARLPGYTVADLNAALYERRSVVKQLAMRRTLWVIRAEDLPA
Mflv_3547|M.gilvum_PYR-GCK PATPPLSLWARLPGFTVADLDAALYEKRSLVKHLAMRRTLWVVRHEDLAA
MSMEG_3806|M.smegmatis_MC2_155 PATPYLSLWARLRRFTVADLDTELYERRTVVKHLAMRRTLWLVGVEDLPF
**** ******* :*****:: ***:*::**:********:: ***.
Mvan_3338|M.vanbaalenii_PYR-1 VQSAAGDRVATNETRRLAADAQNAGVARDGHAWLEAACAAVVRHLRGAGP
Mflv_3547|M.gilvum_PYR-GCK IQAAASDRVAVNEARRLAADVIGAQVAPDGRAWLETASAAVLRHLQERGP
MSMEG_3806|M.smegmatis_MC2_155 IQSAASDRVAANELRKLAADAHNAGLEPDGETWVKVARAAVLRHLEKHGP
:*:**.****.** *:****. .* : **.:*::.* ***:***. **
Mvan_3338|M.vanbaalenii_PYR-1 CTARELREALPELTGTYDPAPGKPYGGEGHLAPRVLTVLSARGLIVRGPN
Mflv_3547|M.gilvum_PYR-GCK TTARELREALPELTGTYDPAPGKPYGGEGHLAPRVLTVLSAQGRIVRGPN
MSMEG_3806|M.smegmatis_MC2_155 TGARELREALPELAGSHDPAPGKSYGGETPLAPRVLTVLGVHGEIMRGPN
***********:*::******.**** *********..:* *:****
Mvan_3338|M.vanbaalenii_PYR-1 DGGWTTSRPRWAAAGSWLG-PADPVSTDQARAELVRRWLHAFGPATVDDL
Mflv_3547|M.gilvum_PYR-GCK DGGWTTSRPRWAAVDRWLP-PAPPVDAEQARADMVGRWLYAFGPATVTDI
MSMEG_3806|M.smegmatis_MC2_155 EGSWTVSRPRWAATADWLGAPYVPAPADEARAELVRRWLATFGPATVTDV
:*.**.*******. ** * *. :::***::* *** :****** *:
Mvan_3338|M.vanbaalenii_PYR-1 KWWFGATLGWARQALSDVDAVEVAVEGSAGTS---------GFVLPGDDG
Mflv_3547|M.gilvum_PYR-GCK KWWFGNTLGSARQALADLGAVEVAIEGSGGSGGAGGSETSVGYVLPGDDE
MSMEG_3806|M.smegmatis_MC2_155 KWWFGHTLTWARHALRDIGAVEVELEGCDAPG----------YVLPDDLD
***** ** **:** *:.**** :**. ... :***.*
Mvan_3338|M.vanbaalenii_PYR-1 PEPDVEPWCALLPGLDVTTMGWAGRDWYLGPHRGAVFDRNGNAGPTVWVD
Mflv_3547|M.gilvum_PYR-GCK PEPAVDSWCALLPGLDVTAMGWYERDWYLGAHRSEIFDRNGNAGPTVWVD
MSMEG_3806|M.smegmatis_MC2_155 PEPEPQPWVALLPGLDVTTMGWFDRDWYLDGLRDQVFDRNGNAGPTVWCD
*** :.* *********:*** *****. *. :************ *
Mvan_3338|M.vanbaalenii_PYR-1 GRVVGAWRQDDDGRVELMLLEDVGRPALRALTARAEELTAWLGGVQVKPR
Mflv_3547|M.gilvum_PYR-GCK GRVVGAWRQDSGGRVELMLLEDVGRRTAGLLAARADEFTAWLGGTRVNPR
MSMEG_3806|M.smegmatis_MC2_155 GRVVGAWAHDADGRVDVRLCTDIGRAARKLLQRKADDLTEWLNGTRVKPR
******* :* .***:: * *:** : * :*:::* **.*.:*:**
Mvan_3338|M.vanbaalenii_PYR-1 FPSPASKSAARR-
Mflv_3547|M.gilvum_PYR-GCK FPSPLSKAGNVRR
MSMEG_3806|M.smegmatis_MC2_155 FPSPLSKGQFTG-
**** **.