For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIAIRDEQESDIAAIRALTLQAFADVPQSRQTEAAVIDALRAAGALTLSLVAVEGGDIVGHVAVSPLSID GDAGPGWYGGGPLSVRPDRQRAGIGTALIRAVFERLRENGARGAVVVGDPRYYERFGFRQATTLALPDVP AENLLVLALDGQCPTGTVAFHPAFGVEDTTPHA*
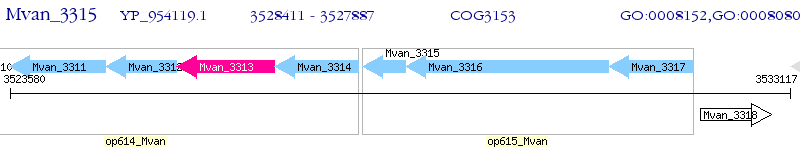
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3315 | - | - | 100% (174) | GCN5-related N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3039c | - | 6e-05 | 27.86% (140) | phosphinothricin N-acetyltransferase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3315|M.vanbaalenii_PYR-1 MSIAIRDEQESDIAAIRALTLQAFADVPQS----RQTEAAVIDALRAAGA
MAB_3039c|M.abscessus_ATCC_199 --MHIRDATAADAAACAAIYAPYVTDTAISFEEVPPSDAVLAERIQTAAH
: *** :* ** *: .:*.. * ::*.: : :::*.
Mvan_3315|M.vanbaalenii_PYR-1 LTLSLVAVEGGDIVGHVAVSPLSIDGDAGPGWYGGGPLSVRPDRQRAGIG
MAB_3039c|M.abscessus_ATCC_199 RHAWLVAEDGGEILGYAYAAPWKSR--TAYQWACEVSVYVDGARRQRGTG
*** :**:*:*:. .:* . :. * .: * *:: * *
Mvan_3315|M.vanbaalenii_PYR-1 TALIRAVFERLRENGARGAVVVGDPRYYERFGFRQATTLALPDVPAENLL
MAB_3039c|M.abscessus_ATCC_199 RQLYTALLGRLRELGYR--TVLAVVVTPNAASEKLHRTMGFEQVALYRRI
* *:: **** * * .*:. : . : *:.: :*. . :
Mvan_3315|M.vanbaalenii_PYR-1 VLALDGQCPTGTVAFHPAFGVEDTTPHA-----
MAB_3039c|M.abscessus_ATCC_199 GYKLGSWHDVTHFQLFLTSAGDSRAPGPAPGEA
*.. . . :. : . :. :* .