For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTAHTASEFRTRLSEADWLDRAAGHRRRADGFLAPHLQRRRAGQAHPVHDFLFTYYSVRPGQLRVWHPGH GVFLDGAAAACYLGRAGYVRRDGGVTVGGDYLHSRMSTVSFIADLLRATASRPARFNCFGMHEWAMVYRA PTVRHSGVPLRLGPDGTDAVVESMPLRCSHFDAYRFFTEPAVDRNAQQLTRQSQITFEQPGCVHAGMDLY KWAVKLGPLIESGLVLDCLELAADARVLDMRASPYDLRAFGFEPIAVETPAGRREYTRAQEAISARAAPL RTRLLQHCEALRDTATAD
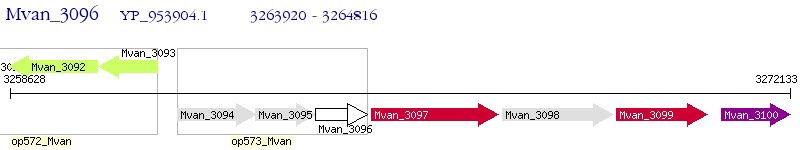
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3096 | - | - | 100% (298) | hypothetical protein Mvan_3096 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3366 | - | 1e-118 | 72.60% (281) | hypothetical protein Mflv_3366 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3639 | - | 1e-100 | 61.30% (292) | hypothetical protein MSMEG_3639 |
| M. thermoresistible (build 8) | TH_0287 | - | 1e-105 | 64.77% (281) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3639|M.smegmatis_MC2_155 MSDAVGTSVTRGHPRRLLDEREWTAAEQRHRERVEEFLAPHVRRMQRREP
TH_0287|M.thermoresistible__bu VSDLCASELS---------EPEWSAREHRHHARVEEFLRAHPHRGRTGRP
Mvan_3096|M.vanbaalenii_PYR-1 MTAHTASEFRTR-----LSEADWLDRAAGHRRRADGFLAPHLQRRRAGQA
Mflv_3366|M.gilvum_PYR-GCK -----MSDSRP-----------WQHLAAAHRDRADDLLAPHVARRQTGQT
:. * *: *.: :* .* * : ..
MSMEG_3639|M.smegmatis_MC2_155 HPVWDFLFTYYSLRPRQLRRWHPGYGIVLTGPSADRFLTYPGYGRHDEGA
TH_0287|M.thermoresistible__bu HPVWDFLFTYYSLRPRQLRRWHPGYGVVLGGAGARRFLDRPGYTAHRG--
Mvan_3096|M.vanbaalenii_PYR-1 HPVHDFLFTYYSVRPGQLRVWHPGHGVFLDGAAAACYLGRAGYVRRDG--
Mflv_3366|M.gilvum_PYR-GCK HPVFDFLFTYYSLKPRRLRVWHPGYGTVLTGPGSERYLDGAGYVQTDA--
*** ********::* :** ****:* .* *..: :* .**
MSMEG_3639|M.smegmatis_MC2_155 GVTVTCEHLASRTDTVRFVAALMRATAARTPRLNCFGLHEWAMVYRTDHV
TH_0287|M.thermoresistible__bu GVTVGAEYLAARIDTVRFIADLLRATADRPPQLNCFGLHEWAMVYRTRTI
Mvan_3096|M.vanbaalenii_PYR-1 GVTVGGDYLHSRMSTVSFIADLLRATASRPARFNCFGMHEWAMVYRAPTV
Mflv_3366|M.gilvum_PYR-GCK GVTVGREYLAARMDTVTFIAGLLGATAARRPRFNCFGLHEWAMVYRTPTV
**** ::* :* .** *:* *: *** * .::****:********: :
MSMEG_3639|M.smegmatis_MC2_155 RHSAVPLRLSAAGTDAVVDSMPLRCTHFDAFRFFTDAAAPRNERQLTRDQ
TH_0287|M.thermoresistible__bu RHADVPLRLGPAGTDAVVEALPLRCTHYDAYRFFTPAAARRNAR-LSRDT
Mvan_3096|M.vanbaalenii_PYR-1 RHSGVPLRLGPDGTDAVVESMPLRCSHFDAYRFFTEPAVDRNAQQLTRQS
Mflv_3366|M.gilvum_PYR-GCK RHARVPLRLGGAGTDEVVESMPLRCSHFDAYRFFTPAAAERNAHRLTRQE
**: *****. *** **:::****:*:**:**** .*. ** : *:*:
MSMEG_3639|M.smegmatis_MC2_155 QIASEQPGCVHASMDLYKWAYKLGPLVASDLVADALELAVDARALDMRAS
TH_0287|M.thermoresistible__bu QRDHEQPGCLHAAMDLYKWAYKLGPLVPSELVLDCLRLSARARALDMRAS
Mvan_3096|M.vanbaalenii_PYR-1 QITFEQPGCVHAGMDLYKWAVKLGPLIESGLVLDCLELAADARVLDMRAS
Mflv_3366|M.gilvum_PYR-GCK QVTAEQPGCVHAGMDLYKWAFKLGPLIESGLVLDCFELAVDARVLDMRAS
* *****:**.******* *****: * ** *.:.*:. **.******
MSMEG_3639|M.smegmatis_MC2_155 PYDLTSYGFEPIAIETPAGRAEYVRAQGQIAERAASIRAALANRCELLLI
TH_0287|M.thermoresistible__bu PYDLRSFGFQPVAIETPAGRAEYVRGQQEIARRAATLRSVLLDRCEALLA
Mvan_3096|M.vanbaalenii_PYR-1 PYDLRAFGFEPIAVETPAGRREYTRAQEAISARAAPLRTRLLQHCEALRD
Mflv_3366|M.gilvum_PYR-GCK PYDLRAYGFEPIAVETADGRREYARAQEQISERAAPLRTRLIDRCRALQA
**** ::**:*:*:**. ** **.*.* *: ***.:*: * ::*. *
MSMEG_3639|M.smegmatis_MC2_155 DMTVETPTLPMGKLDKRQAPPGAEEPREESMTTHRAAKAVRA
TH_0287|M.thermoresistible__bu AAADSTVCAR--------------------------------
Mvan_3096|M.vanbaalenii_PYR-1 TATAD-------------------------------------
Mflv_3366|M.gilvum_PYR-GCK ADTGE-------------------------------------
: .