For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTTSTRTGRKRFTLIDAGREFWRHPSPWMIAATLVVASGARIAYGDWQITDALVPAIMLAAFPFFEWLV HVCILHWRPRKIGVLRIDPLLARKHREHHVEPREIALIFIPWQALLWVLPVALGIALLAFPRTALGLTFL TFLTVLGLCYEWCHYLVHTDYKPKTAAYRAVWRNHRQHHFKNEHYWFTVTSAGTADRVLGTYPDPATVPT SPTAKNLHGQA
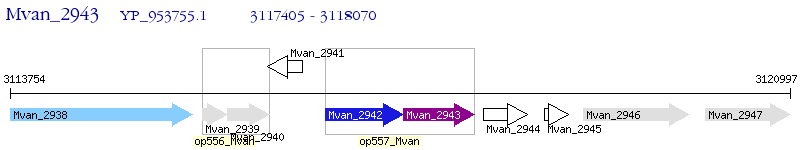
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2943 | - | - | 100% (221) | fatty acid hydroxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3019 | - | 9e-73 | 56.42% (218) | fatty acid hydroxylase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3529 | - | 1e-93 | 70.33% (209) | fatty acid hydroxylase |
| M. thermoresistible (build 8) | TH_1960 | - | 5e-94 | 72.73% (209) | fatty acid hydroxylase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2943|M.vanbaalenii_PYR-1 MTTTSTRTGRKRFTLIDAGREFWRHPSPWMIAATLVVASGARIAYGDWQI
TH_1960|M.thermoresistible__bu -------VARRGFTLADAFREFRRHPSPWLIAGTLVTAAAARAVLGDWQL
MSMEG_3529|M.smegmatis_MC2_155 -------MSRRDFTLADAGREFVRHPSPWMIGVTLAAAATARTIVGDWQV
MAB_3019|M.abscessus_ATCC_1997 ---MTRTATRKGMTLRDALAEFIKHPTPWMLLAWSSALIIGRITLGSWSI
*: :** ** ** :**:**:: . .* *.*.:
Mvan_2943|M.vanbaalenii_PYR-1 TDALVPAIMLAAFPFFEWLVHVCILHWRPRKIGVLRIDPLLARKHREHHV
TH_1960|M.thermoresistible__bu TDALVPVVMLALFPFLEWVVHVGILHWRPRRLGSLIVDWRLARKHREHHQ
MSMEG_3529|M.smegmatis_MC2_155 TDAVVPLVMVGVFPLAEWVIHVAILHWRPRRLGPVVVDTRLARDHRRHHA
MAB_3019|M.abscessus_ATCC_1997 ADAIVPIALVAISPIAEWLIHVGILHWRPRSLGPVTLDSRLARDHRLHHQ
:**:** ::. *: **::** ******* :* : :* ***.** **
Mvan_2943|M.vanbaalenii_PYR-1 EPREIALIFIPWQALLWVLPVALGIALLAFPRTALGLTFLTFLTVLGLCY
TH_1960|M.thermoresistible__bu DPRDVPLIFIPWQSLIWVLILAVAVALLVFPRPALGVTFLTCLTVLGLGY
MSMEG_3529|M.smegmatis_MC2_155 EPRDVPLIFIPWPTLLWLLPVATAVALVAFPRPGLGLTFLTLLTALGLGY
MAB_3019|M.abscessus_ATCC_1997 DPRDVPLVFIPWPSLIVVIVGVTAIGLLMFPRNGLGLTFALAIALFLMFY
:**::.*:**** :*: :: . .:.*: *** .**:** :: : : *
Mvan_2943|M.vanbaalenii_PYR-1 EWCHYLVHTDYKPKTAAYRAVWRNHRQHHFKNEHYWFTVTSAGTADRVLG
TH_1960|M.thermoresistible__bu EWCHYLIHSDYKPKSRWYRAIWRNHRQHHYKNERYWFTVTSAQTADRVLG
MSMEG_3529|M.smegmatis_MC2_155 EWCHYLIHSDYKPRSDVYRAIWRNHRRHHFKNEHYWFTVTSAGTADRVLG
MAB_3019|M.abscessus_ATCC_1997 EWTHYLIHTDYKPRRAIYRAVWRNHRYHHFKNENYWFTVTSSGTADRLLG
** ***:*:****: ***:***** **:***.*******: ****:**
Mvan_2943|M.vanbaalenii_PYR-1 TYPDPATVPTSPTAKNLHGQA---
TH_1960|M.thermoresistible__bu TCPDPATAPTSPTARTLHA-----
MSMEG_3529|M.smegmatis_MC2_155 TCPDPATVPTSPTARQLHVTPAR-
MAB_3019|M.abscessus_ATCC_1997 TFPDPQQVKSSPTVRNLHAPSITG
* *** . :***.: **