For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRVPVVLSTVATIALCGVSWGAPPATADNADWGLNGTYIATSNGEWARKNEVFYDQASLRSTWTVSTQCS YPGECTGTVDSEWGWTAPIYQKSGVWYVKHTVENWVPCPDGTTAPGLQVFRFKAMNAEGAMADPTSDTLV GEDITTGVSGACGVSKPVYINMPFKLVKVG*
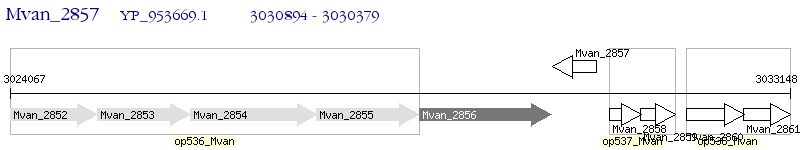
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2857 | - | - | 100% (171) | hypothetical protein Mvan_2857 |
| M. vanbaalenii PYR-1 | Mvan_4144 | - | 3e-28 | 38.95% (172) | hypothetical protein Mvan_4144 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4532 | - | 9e-92 | 86.55% (171) | hypothetical protein Mflv_4532 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4704 | - | 2e-34 | 44.72% (161) | hypothetical protein MMAR_4704 |
| M. avium 104 | MAV_0954 | - | 3e-31 | 39.64% (169) | hypothetical protein MAV_0954 |
| M. smegmatis MC2 155 | MSMEG_2861 | - | 6e-76 | 76.92% (169) | hypothetical protein MSMEG_2861 |
| M. thermoresistible (build 8) | TH_2439 | - | 3e-79 | 78.24% (170) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2857|M.vanbaalenii_PYR-1 --MTRVP-VVLSTVATIALCGVSWGAPPATADNA-DWGLNGTYIATSNGE
Mflv_4532|M.gilvum_PYR-GCK --MIRVP-SAAGAVAAVVICGAAWGAPPAAADNP-EWGLNGTYIATSNGE
TH_2439|M.thermoresistible__bu VAMVRIP-VVLAAVTALGLA----TSPDAGADTA-DWGLNGTYIATSNGE
MSMEG_2861|M.smegmatis_MC2_155 --MMRVSRVSIVMATAAATCFGVATSAPAAAEN--DWGLNGTYVATSNGE
MMAR_4704|M.marinum_M --MRSMGVAISATLI----LGGVGSAPSAWAY---DPAINGTYTATVIGD
MAV_0954|M.avium_104 --MRSVRTLTTATLVAATAFGGLGAAATAGATTKEEVAVNGTFRATSIGD
* : :. * * : .:***: ** *:
Mvan_2857|M.vanbaalenii_PYR-1 WARKNEVFYDQASLRSTWTVSTQCSYPGECTGTVDSEWGWTAPIYQKSG-
Mflv_4532|M.gilvum_PYR-GCK WARKNEVFYDQASLRSTWTVSTQCSYPGECTGTVDSEWGWSAPIYQKSG-
TH_2439|M.thermoresistible__bu LARKNEVFFEQRSLRSTWTISTQCSYPGECTGTVVSEWGWTAPIYQRSG-
MSMEG_2861|M.smegmatis_MC2_155 WARKNDVFYEQASLRSTWTISTQCSYPGECTGTVHSEWGWTAPIYQKSG-
MMAR_4704|M.marinum_M WARTNTVYHDEPVVRSTWTITSSCSTAQDCSGQVTSDQGWTAPLTMHDGQ
MAV_0954|M.avium_104 WAQRNDQYFEEPTVVQTWTISSTCDTFQECHGTVTSDLGWTAPLYMSDGQ
*: * :.:: : .***::: *. :* * * *: **:**: .*
Mvan_2857|M.vanbaalenii_PYR-1 VWYVKHTVENWVPCPDGTTAPGLQVFRFKAMNAEGAMADPTSDTLVGEDI
Mflv_4532|M.gilvum_PYR-GCK VWYVKHTVENWIPCPDGTSAPGLQVFRFKAMNAEGAMADPTATTLVGEDV
TH_2439|M.thermoresistible__bu VWYVKHTIEDWIPCPDGSTAPGLQVFRFKAMTPDGTTADPTATTLVGEDI
MSMEG_2861|M.smegmatis_MC2_155 IWYVKHTIPDWVPCPDGTAAPGLQVFRFKPMTPNGAMNDPKSTTLVGEDI
MMAR_4704|M.marinum_M LWTVKRDLPNWETCPDGTSFTGHDVIYFYPANPDTGETQLGSPVLAGREK
MAV_0954|M.avium_104 MWKVKHEVPNWERCEDGTAFTGQQTFYFYPVN-EYGGYQLGSSTFAGKDK
:* **: : :* * **:: .* :.: * . . : : : .:.*.:
Mvan_2857|M.vanbaalenii_PYR-1 TTGVSGACGVSKPVYINMPFKLVKVG
Mflv_4532|M.gilvum_PYR-GCK TTGVSGACGVSRSVYINMPFKLVKVG
TH_2439|M.thermoresistible__bu TTGVSGACGVNKPVYINMPFKLVKV-
MSMEG_2861|M.smegmatis_MC2_155 TTGESGACGVNKPVYITMPFKLVKT-
MMAR_4704|M.marinum_M TTGPSGACGNNAPLYVEQPFRLDKIG
MAV_0954|M.avium_104 TVGPSGACGQNQWLDIAMPLRLDKLS
*.* ***** . : : *::* *