For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IVELADTTTNEINKKIIGLREEGGALTLGRVLTLVIVPDCEETLEDAIEAATFASREHPCRVIVTLSGDR DAAQSRLDAELRVGRDAGAGEVVVLRLHGELADHAESVVLPFLLADTPVVAWWPAAAPAIPSQDPVGRLA IRRITDATSCADPLAAIQSRLPAYSPGDTDISWSRITYWRALLASAIDDPPHEPITSAVVSGLQSEPALD LLAGWLASRIDGAVTRAVGTLKVELHRGSETVTLSRPQDGRTATLSRTSRPVALVPLARRDVPECLAEEL RRLDADEIYHEALSGIDKVTYR*
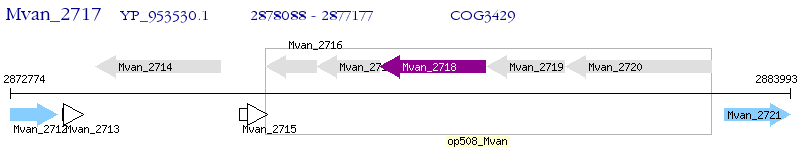
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2717 | - | - | 100% (303) | OpcA protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1481c | opcA | 1e-117 | 69.64% (303) | putative OXPP cycle protein OPCA |
| M. gilvum PYR-GCK | Mflv_3695 | - | 1e-145 | 84.16% (303) | OpcA protein |
| M. tuberculosis H37Rv | Rv1446c | opcA | 1e-116 | 69.31% (303) | putative OXPP cycle protein OPCA |
| M. leprae Br4923 | MLBr_00580 | - | 3e-67 | 56.07% (239) | hypothetical protein MLBr_00580 |
| M. abscessus ATCC 19977 | MAB_2762 | - | 1e-112 | 66.56% (302) | putative OxpP cycle protein OpcA |
| M. marinum M | MMAR_2251 | opcA | 1e-120 | 70.86% (302) | OXPP cycle protein OpcA |
| M. avium 104 | MAV_3330 | opcA | 1e-116 | 68.54% (302) | OpcA protein |
| M. smegmatis MC2 155 | MSMEG_3100 | opcA | 1e-135 | 78.48% (302) | OpcA protein |
| M. thermoresistible (build 8) | TH_0336 | opcA | 1e-121 | 69.21% (302) | PUTATIVE OXPP CYCLE PROTEIN OPCA |
| M. ulcerans Agy99 | MUL_1841 | opcA | 1e-119 | 70.20% (302) | OXPP cycle protein OpcA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1481c|M.bovis_AF2122/97 ----MIVDLPDTTTTAVNKKLDELREKIGAVAMGRVLTLIIAPDSEAMLE
Rv1446c|M.tuberculosis_H37Rv ----MIVDLPDTTTTAVNKKLDELREKIGAVAMGRVLTLIIAPDSEAMLE
MMAR_2251|M.marinum_M ----MIVDLPDTTTTAVNKKLDELREKIGAVTMGRVLTLIIVPDSETMVE
MUL_1841|M.ulcerans_Agy99 ----MIVDLPDTTTTAVNKKLDELREKIGAVTMGRVLTLIIVPDSETMVE
MAV_3330|M.avium_104 -MAQLIIDMPDATTTAVNKKLNELRERVGAVAMGRVLTLIITPDSEEILE
Mvan_2717|M.vanbaalenii_PYR-1 ----MIVELADTTTNEINKKIIGLREEGGALTLGRVLTLVIVPDCEETLE
Mflv_3695|M.gilvum_PYR-GCK ----MIVELSDTTTNEINKKITGLREEGGALTLGRVLTLVITPDCEENLA
MSMEG_3100|M.smegmatis_MC2_155 ----MIVDLPDTSTNDVNKKITGLREEGGALTLSRVLTLVIAPHTEDLVE
TH_0336|M.thermoresistible__bu ----LIVDLPDTTTNAINKKITALREQAGAITLGRVLTLVVAADSEDQLD
MAB_2762|M.abscessus_ATCC_1997 MEAAMIIDLPNTTTNDVNKKLVELRETGGAVTMARVLTLVVITDAGDDVE
MLBr_00580|M.leprae_Br4923 --------------------------------MSRVLTLVITPYSKAMLK
:.*****:: . :
Mb1481c|M.bovis_AF2122/97 ESIEAANDASHEHPSRIIVTMRGDPYADRPRLDAQLRVGADAGAGEFVVL
Rv1446c|M.tuberculosis_H37Rv ESIEAANDASHEHPSRIIVTMRGDPYADRPRLDAQLRVGADAGAGEFVVL
MMAR_2251|M.marinum_M ESIDAANDASHEHPSRIIVTMRGDPYADEPRLDAQLRAGGDAGSGEVVVL
MUL_1841|M.ulcerans_Agy99 ESIDAANDASHEHPSRIIVTMRGDPYADEPRLDAQLRAGGDPGSGEVVVL
MAV_3330|M.avium_104 ESLKAANDASHEHPSRIIVTLRGNPYADKPRLDAQLRAGGDTGASEVVVL
Mvan_2717|M.vanbaalenii_PYR-1 DAIEAATFASREHPCRVIVTLSGDRDAAQSRLDAELRVGRDAGAGEVVVL
Mflv_3695|M.gilvum_PYR-GCK SAIEAANFASREHPCRVIVALQADR-SAAARLDAELRVGRDAGAGEVVVL
MSMEG_3100|M.smegmatis_MC2_155 DSIEAANFASREHPCRVIVVVPGDRDAAAPRLDAQLRVGHDAGAGEVVVL
TH_0336|M.thermoresistible__bu DAIAAATSASREHPCRIIVVVPVDRSTPEPRLDGQLRVGGDAGVGEVVVL
MAB_2762|M.abscessus_ATCC_1997 EVIEAANGASHEHPCRVIVLE-RNPLASDTGLSAQIRVGGDAGTGEVVVL
MLBr_00580|M.leprae_Br4923 ESIEAANGASHKYPNRIIIAMRVNSYANKARLDAQLWVGADTGAG-VVVS
. : **. **:::* *:*: : : . *..:: .* *.* . .**
Mb1481c|M.bovis_AF2122/97 RLSGPLAGHADSVVIPFLLPDIPVVAWWPDIAPAVPAQDALGKLAIRRIT
Rv1446c|M.tuberculosis_H37Rv RLSGPLAGHADSVVIPFLLPDIPVVAWWPDIAPAVPAQDALGKLAIRRIT
MMAR_2251|M.marinum_M RLSGPLAGHAESVVTPFLLPDIPVVAWWPDIAPAVPAQDPLGKLAIRRIT
MUL_1841|M.ulcerans_Agy99 RLSGPLAGHAESVVTPFLLPDIPVVAWWPDIAPAVPTQDPLGKLAIRRIT
MAV_3330|M.avium_104 WLSGALSGHAASVVTPFLLPDIPVVVWWPDVAPAVPAQDPLGRLAIRRIT
Mvan_2717|M.vanbaalenii_PYR-1 RLHGELADHAESVVLPFLLADTPVVAWWPAAAPAIPSQDPVGRLAIRRIT
Mflv_3695|M.gilvum_PYR-GCK RLYGELADHADSVVLPFLLPDTPVVAWWPASAPETPAEDPVGKLAIRRIT
MSMEG_3100|M.smegmatis_MC2_155 RLSGELADHSASVVLPFLLPDTPVVAWWPAVAPAVPSEDPLGKLAIRRIT
TH_0336|M.thermoresistible__bu RIAGPLADQAASVVLPFLLPDTPVVAWWPGEAPPVPAQDPLGRLAVKRIT
MAB_2762|M.abscessus_ATCC_1997 RLRGPLAAHEHSVVIPFLLPDTPVVAWWPNQAPPIPAQHPLGRLAIRRIT
MLBr_00580|M.leprae_Br4923 RT---LAVYAHSVVISILLPDIPMVAWWPNIAPTMSGQDSLGKLAIQRIT
*: *** .:**.* *:*.*** ** . :..:*:**::***
Mb1481c|M.bovis_AF2122/97 DATNAIDPLSAIKSRLAGYGAGDTDLAWSRITYWRALLTSAVDQPPHEPI
Rv1446c|M.tuberculosis_H37Rv DATNAIDPLSAIKSRLAGYGAGDTDLAWSRITYWRALLTSAVDQPRHEPI
MMAR_2251|M.marinum_M DATNGIDPLSAIKSRLPGYTAGDTDLAWSRITYWRALLTSAVDQPPHEPI
MUL_1841|M.ulcerans_Agy99 DATNGIDPLSAIKSRLPGYTAGDTDLAWSRITYWRALLTSAVDQPPHEPI
MAV_3330|M.avium_104 DATNGVDPLAAIKSRLPGYTAGDTDLAWARITYWRALLAAAVDLAPHEPI
Mvan_2717|M.vanbaalenii_PYR-1 DATSCADPLAAIQSRLPAYSPGDTDISWSRITYWRALLASAIDDPPHEPI
Mflv_3695|M.gilvum_PYR-GCK DATLASDPLAAIKSRLPGYSPGDTDIAWSRITYWRALLASAIDEPPHEPI
MSMEG_3100|M.smegmatis_MC2_155 DATNGTDPLACIKSRLKAYSPGDTDLAWSRVTYWRALLAAAVDQEPFEPV
TH_0336|M.thermoresistible__bu DATASSDPLTAIKGRLRGYTPGDTDLAWSRVTYWRALLASAVDQPPYEPI
MAB_2762|M.abscessus_ATCC_1997 DATTAPDPMNAIKNRLAGYTPGDTDLSWSRITYWRALLASALDQAPYEGI
MLBr_00580|M.leprae_Br4923 NATNSIDPLATIKSRLSDYTADDTHLAWDLITYWRALLTSAVNLPPHEPI
:** **: *:.** * ..**.::* :*******::*:: .* :
Mb1481c|M.bovis_AF2122/97 ESALVSGLKTEPALDVLAGWLASRIEGPVRRAVGELKVELVRNSETIVLS
Rv1446c|M.tuberculosis_H37Rv ESALVSGLKTEPALDVLAGWLASRIEGPVRRAVGELKVELVRNSETIVLS
MMAR_2251|M.marinum_M VSALVSGLRTEPALDILAGWLASRIDGPVRRAVGELRVELERPSENIVLS
MUL_1841|M.ulcerans_Agy99 VSALVSGLRTEPALDILAGWLASRIDGPARRAVGELRVELERPSENIVLS
MAV_3330|M.avium_104 ESALVSGLKTEPALDVLAGWLASRIDGPVRRAVGELKVELARSSETIVLS
Mvan_2717|M.vanbaalenii_PYR-1 TSAVVSGLQSEPALDLLAGWLASRIDGAVTRAVGTLKVELHRGSETVTLS
Mflv_3695|M.gilvum_PYR-GCK ERAVVSGLKAEPALDILAGWLASRIDGEVTRVVGTLKVELHRASETVTLS
MSMEG_3100|M.smegmatis_MC2_155 SSAVVSGLKDEPALDILAGWLASRIDGPVTRAVGELKVELNRPSETVTLS
TH_0336|M.thermoresistible__bu ESALVSGLATEPALDVLAGWLASRLDCEVRRATGPLQVQLVRSSETITLG
MAB_2762|M.abscessus_ATCC_1997 NSAVVSGLATEPALDVTAGWLASRIDGPVTRVVGDLKVELHRDSEIITLS
MLBr_00580|M.leprae_Br4923 DLALVSGMKTEPALDVLAGWLANRINRPLRRAVADLKVELIRNSETIVLS
*:***: *****: *****.*:: *... *:*:* * ** :.*.
Mb1481c|M.bovis_AF2122/97 RPQEGITATLTRTGKPDALVPLARRVTGECLAEDLRRLDPDEIYCAALEG
Rv1446c|M.tuberculosis_H37Rv RPQEGITATLTRTGKPDALVPLARRVTGECLAEDLRRLDPDEIYCAALEG
MMAR_2251|M.marinum_M RPQEGITATLSRTAKPDALVPLARRVVGECLAEDLRRLDADEIYFAALEG
MUL_1841|M.ulcerans_Agy99 RPQEGITATLSRTAKPDALVPLARRVVGECLAEDLRRLDADEIYFAALEG
MAV_3330|M.avium_104 RPQEGRTATLSRTSRPDALLPLARRETGECLAEDLRRLDADEIYQSALEG
Mvan_2717|M.vanbaalenii_PYR-1 RPQDGRTATLSRTSRPVALVPLARRDVPECLAEELRRLDADEIYHEALSG
Mflv_3695|M.gilvum_PYR-GCK RPQEGLTATLSRTSRPVALIPLARRDIPDCLAEELRRLDADEIYREALTG
MSMEG_3100|M.smegmatis_MC2_155 RPQDGITATLSRTGKPAALVPLARREAKECLAEDLRRLDADEIYREALQG
TH_0336|M.thermoresistible__bu RPQDGVTGTLSRTGKPDALLPLGRRETRECLAEDLRRLDPDEIYHEALRG
MAB_2762|M.abscessus_ATCC_1997 RPQTGAIATLTRTGRPSASLPLPRRETRDCLAEDLRRLDPDEIYRTALEG
MLBr_00580|M.leprae_Br4923 RPQTWVTSTLIRTVKPDALVPWGAQGSRGVPSRKSATTGSRQVLLQCLRR
*** .** ** :* * :* : :.. .. :: .*
Mb1481c|M.bovis_AF2122/97 IKKVQYR
Rv1446c|M.tuberculosis_H37Rv IKKVQYR
MMAR_2251|M.marinum_M IKKVQYV
MUL_1841|M.ulcerans_Agy99 IRKVQYV
MAV_3330|M.avium_104 IEKVQYV
Mvan_2717|M.vanbaalenii_PYR-1 IDKVTYR
Mflv_3695|M.gilvum_PYR-GCK LDKVIYQ
MSMEG_3100|M.smegmatis_MC2_155 IDKVQYV
TH_0336|M.thermoresistible__bu LDKVLYI
MAB_2762|M.abscessus_ATCC_1997 IAKVQYV
MLBr_00580|M.leprae_Br4923 H------