For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGIKSLDDLLSEGVTGRGVLVRSDLNVPVDDDGNISDPGRIIASVPTLKALSDAGAKVVVTAHLGRPKGE PDPKFSLAPVAKALGEKLGRHVQLAGDVVGTDALARAEGLTDGDILLLENIRFDPRETSKDDAERLKLAK ALVELVGGEDGSSAAFVSDGFGVVHRKQASVYDVATLLPHYAGTLVDTEIKVLAQLTEAGERPYAVVLGG SKVSDKLAVIENLATKADSLVIGGGMCFTFLAAQGLSVGSSLLEESMVETCRKLLDDYGDVLHLPVDIVV AEKFAADSPAETVAADRIPDDKMGLDIGPESVKRFTALLSNAKTIFWNGPMGVFEFPAFAAGTKGVAEAI IGATGKGAFSVVGGGDSAAAVRQLGLPEDGFSHISTGGGASLEYLEGKELPGIQVLE
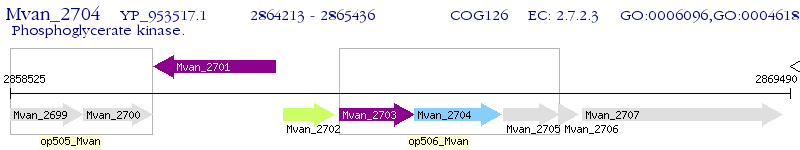
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2704 | pgk | - | 100% (407) | phosphoglycerate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1472 | pgk | 1e-176 | 77.34% (406) | phosphoglycerate kinase |
| M. gilvum PYR-GCK | Mflv_3709 | pgk | 0.0 | 92.36% (406) | phosphoglycerate kinase |
| M. tuberculosis H37Rv | Rv1437 | pgk | 1e-176 | 77.34% (406) | phosphoglycerate kinase |
| M. leprae Br4923 | MLBr_00571 | pgk | 1e-170 | 75.74% (408) | phosphoglycerate kinase |
| M. abscessus ATCC 19977 | MAB_2778c | - | 0.0 | 81.02% (411) | phosphoglycerate kinase (PGK) |
| M. marinum M | MMAR_2240 | pgk | 0.0 | 80.69% (404) | phosphoglycerate kinase Pgk |
| M. avium 104 | MAV_3340 | pgk | 0.0 | 80.44% (409) | phosphoglycerate kinase |
| M. smegmatis MC2 155 | MSMEG_3085 | pgk | 0.0 | 87.71% (407) | phosphoglycerate kinase |
| M. thermoresistible (build 8) | TH_0427 | pgk | 0.0 | 83.29% (407) | PROBABLE PHOSPHOGLYCERATE KINASE PGK |
| M. ulcerans Agy99 | MUL_1829 | pgk | 0.0 | 80.20% (404) | phosphoglycerate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2704|M.vanbaalenii_PYR-1 MGIKSLDDLLSEGVTGRGVLVRSDLNVPVDDD----GNISDPGRIIASVP
Mflv_3709|M.gilvum_PYR-GCK MGIKSLDDLLAEGVSGRGVLVRSDLNVPLDG-----GSITDPGRIIASVP
MSMEG_3085|M.smegmatis_MC2_155 MSVKTLDDLLAEGVQGRGVLVRSDLNVPLDDD----GNITDPGRVIASVP
TH_0427|M.thermoresistible__bu MTVKTLEDLLAEGVSGRGVLVRSDLNVPLDDS----GDITDPGRVIASVP
MAB_2778c|M.abscessus_ATCC_199 MAIKSLNDLLAEGVSGKGVLVRSDLNVPLEYLDGNIAHISDPGRIVASVP
Mb1472|M.bovis_AF2122/97 MSVANLKDLLAEGVSGRGVLVRSDLNVPLDEDG----TITDAGRIIASAP
Rv1437|M.tuberculosis_H37Rv MSVANLKDLLAEGVSGRGVLVRSDLNVPLDEDG----TITDAGRIIASAP
MLBr_00571|M.leprae_Br4923 MRIPNLKDLLEEGVSGRCVLVRCDLNVPLGDDG----AITDLGRVTASVP
MMAR_2240|M.marinum_M MTIHSLEDLLAEGVSGRGVLVRSDLNVPLDENG----VITDAGRITASVP
MUL_1829|M.ulcerans_Agy99 MTIHSLEDLPAEGVSGRGVLVRSDLNVPLDENG----VITDAGRITASVP
MAV_3340|M.avium_104 MAVHNLKDLLAEGVSGRGVLVRSDLNVPLDSDGEQ-GRITDPGRITASVP
* : .*.** *** *: ****.*****: *:* **: **.*
Mvan_2704|M.vanbaalenii_PYR-1 TLKALSDAGAKVVVTAHLGRPKGEPDPKFSLAPVAKALGEKLGRHVQLAG
Mflv_3709|M.gilvum_PYR-GCK TLKALSDAGAKVVVTAHLGRPKGEPDPKFSLAPVGAALGEKLGRHVQLAG
MSMEG_3085|M.smegmatis_MC2_155 TLQALAEAGAKVIVTAHLGRPKGEPDPKLSLAPVAAALGEKLGRHVQLAG
TH_0427|M.thermoresistible__bu TLKALAEAGAKVVVTAHLGRPKGTPDPKLSLAPVAAALGERLGRHVQLAG
MAB_2778c|M.abscessus_ATCC_199 TIRALAGAGAKVIVTAHLGRPDGKPDPKLSLAPVGAALGELLGQHVQVAG
Mb1472|M.bovis_AF2122/97 TLKALLDADAKVVVAAHLGRPKDGPDPTLSLAPVAVALGEQLGRHVQLAG
Rv1437|M.tuberculosis_H37Rv TLKALLDADAKVVVAAHLGRPKDGPDPTLSLAPVAVALGEQLGRHVQLAG
MLBr_00571|M.leprae_Br4923 TLKALLEAGAKIVVAAHLGRPKNGPDPKLSLEPVAAALGEQLGQNVQLVC
MMAR_2240|M.marinum_M TLKALLDGGAKVVVAAHLGRPKDGPDPKLSLEPVAAALGEQLGRHVQLAG
MUL_1829|M.ulcerans_Agy99 TLKALLDGGAKVVVAAHLGRPKDGPDPKLSLEPVAAALGEQLGRHVQLAG
MAV_3340|M.avium_104 TLSALVEAGAKVVVAAHLGRPKNGPDPALSLAPVAAALGEQLGRHVQLAS
*: ** ..**::*:******.. *** :** **. **** **::**:.
Mvan_2704|M.vanbaalenii_PYR-1 ----DVVGTDALARAEGLTDGDILLLENIRFDPRETSKDDAERLKLAKAL
Mflv_3709|M.gilvum_PYR-GCK ----DVVGSDALARAEGLTDGDILLLENIRFDGRETSKDDAERLKLAKAL
MSMEG_3085|M.smegmatis_MC2_155 ----DVVGTDALARAEGLTDGDVLLLENIRFDARETSKDDSERLSLAKAL
TH_0427|M.thermoresistible__bu ----DVVGADALARAEGLTDGDVLLLENVRFDPRETSKDDGERAALARAL
MAB_2778c|M.abscessus_ATCC_199 ----DVVGTDALARAEGLTDGDVLLLENIRFDPRETSKDDAQRLALAREL
Mb1472|M.bovis_AF2122/97 ----DVVGADALARAEGLTGGDILLLENIRFDKRETSKNDDDRRALAKQL
Rv1437|M.tuberculosis_H37Rv ----DVVGADALARAEGLTGGDILLLENIRFDKRETSKNDDDRRALAKQL
MLBr_00571|M.leprae_Br4923 STDRSPVGGDALACVERLTDGDLLLLQNIRFDPRETSKVDDERLALAKQL
MMAR_2240|M.marinum_M ----DIVGTDALARAEGLTDGDVLLLENIRFDKRETSKDDGERLALAKQL
MUL_1829|M.ulcerans_Agy99 ----DIVGTDALARAEGLTDGDVLLLENIRFDKRETSKDDGERLAFAKQL
MAV_3340|M.avium_104 ----DVVGTDALARAEGLTDGDVLLLENIRFDARETSKDDAERLALARQL
. ** **** .* **.**:***:*:*** ***** * :* :*: *
Mvan_2704|M.vanbaalenii_PYR-1 VELVGGEDGSSAAFVSDGFGVVHRKQASVYDVATLLPHYAGTLVDTEIKV
Mflv_3709|M.gilvum_PYR-GCK VELVG-DDG---AFVSDGFGVVHRKQASVYDVATLLPHYAGTLVDTEVKV
MSMEG_3085|M.smegmatis_MC2_155 AALVEGPDGSPGVFVSDGFGVVHRKQASVYDVATLLPHYAGTLVAAEVKV
TH_0427|M.thermoresistible__bu VELVG-DDA---AFVSDGFGVVHRKQASVYDVAKLLPAYAGTLVATEVKV
MAB_2778c|M.abscessus_ATCC_199 AELVGGPTGTGGAFVSDGFGVVHRKQASVYDVATLLPHYAGGLVAAEVEV
Mb1472|M.bovis_AF2122/97 VELVG----TGGVFVSDGFGVVHRKQASVYDIATLLPHYAGTLVADEMRV
Rv1437|M.tuberculosis_H37Rv VELVG----TGGVFVSDGFGVVHRKQASVYDIATLLPHYAGTLVADEMRV
MLBr_00571|M.leprae_Br4923 VELVG----SAGAFVSDGFGVVHRRQASVYDVATLLPHYAGILVADEIRI
MMAR_2240|M.marinum_M AELVS----PGGAFVSDGFGVVHRKQASVYDVATLLPHYAGRLVADEIRV
MUL_1829|M.ulcerans_Agy99 AELVS----PEGAFVSDGFGVVHRKQASVYDVATLLPHYAGRLVADEIRV
MAV_3340|M.avium_104 AELVG----PTGAFVSDGFGVVHRKQASVYDVATLLPHYAGTLVAEEIAV
. ** .***********:******:*.*** *** ** *: :
Mvan_2704|M.vanbaalenii_PYR-1 LAQLTEAGERPYAVVLGGSKVSDKLAVIENLATKADSLVIGGGMCFTFLA
Mflv_3709|M.gilvum_PYR-GCK LAQLTESTERPYAVVLGGSKVSDKLAVIENLATKADSLIIGGGMCFTFLA
MSMEG_3085|M.smegmatis_MC2_155 LQQLTSSTDRPYAVVLGGSKVSDKLAVIENLATKADSLIIGGGMCFTFLA
TH_0427|M.thermoresistible__bu LEQLTESTDRPYAVVLGGSKVSDKLAVIERLAQKADSLVIGGGMCFTFLA
MAB_2778c|M.abscessus_ATCC_199 LRVLTASTERPYAVVLGGSKVSDKLAVIESLATKADSLVIGGGMCFTFLA
Mb1472|M.bovis_AF2122/97 LEQLTSSTQRPYAVVLGGSKVSDKLGVIESLATKADSIVIGGGMCFTFLA
Rv1437|M.tuberculosis_H37Rv LEQLTSSTQRPYAVVLGGSKVSDKLGVIESLATKADSIVIGGGMCFTFLA
MLBr_00571|M.leprae_Br4923 LEQLTSSAKRPYAVVLGGSKVSDKLGVIESLATKADSIVLGGGMCFTFLA
MMAR_2240|M.marinum_M LEQLTSSTERPYAVVLGGSKVSDKLGVIESLATKADSIVIGGGMCFTFLA
MUL_1829|M.ulcerans_Agy99 LEQLTSSTERPYAVVLGGSKVSDKLGVIESLATKADSIVIGGGMCFTFLA
MAV_3340|M.avium_104 LEQLTGSTKRPYAVVLGGSKVSDKLGVIESLATKADSIVIGGGMCFTFLA
* ** : .****************.*** ** ****:::**********
Mvan_2704|M.vanbaalenii_PYR-1 AQGLSVGSSLLEESMVETCRKLLDDYGDVLHLPVDIVVAEKFAADSPAET
Mflv_3709|M.gilvum_PYR-GCK AQGLSVGSSLLEESMIDTCRTLLDDYGDVLHLPVDIVVAEKFSADSEPQT
MSMEG_3085|M.smegmatis_MC2_155 AQGFSVGSSLLQEEMVDTCRRLLDEYADVIHLPVDIVVADKFAADAEAET
TH_0427|M.thermoresistible__bu SQGLSVGKSLVQPEMIDTCRQLLDTYADVIHVPVDIVVADEFAADSPSEI
MAB_2778c|M.abscessus_ATCC_199 AQGLPVGKSLVQLEMVDTCKRLLDTYADVIHLPMDIVAADEFAADSPSEI
Mb1472|M.bovis_AF2122/97 AQGFSVGTSLLEDDMIEVCRGLLETYHDVLRLPVDLVVTEKFAADSPPQT
Rv1437|M.tuberculosis_H37Rv AQGFSVGTSLLEDDMIEVCRGLLETYHDVLRLPVDLVVTEKFAADSPPQT
MLBr_00571|M.leprae_Br4923 AQGFSVGKSLLETEMIETCRSLLDTYADVLRLPMDIVVTEKFVADSPPQT
MMAR_2240|M.marinum_M AQGYSVGTSLLEKEMIDTCRRLLDTYHDVLRLPVDVVVTEKFAADSPPQT
MUL_1829|M.ulcerans_Agy99 AQGYSVGTSLLEKEMIDTCRRLLDTYHDVLRLPVDVVVTEKFAADPPPQT
MAV_3340|M.avium_104 AQGFSVGKSLLETEMVDTCRRLLDTYVDVLRLPVDIVAADRFAADAAPQT
:** .**.**:: .*::.*: **: * **:::*:*:*.::.* **. .:
Mvan_2704|M.vanbaalenii_PYR-1 VAADRIPDDKMGLDIGPESVKRFTALLSNAKTIFWNGPMGVFEFPAFAAG
Mflv_3709|M.gilvum_PYR-GCK VAADKIPDDKMGLDIGPESVRRFSALLSNAKTIFWNGPMGVFEFPAFAAG
MSMEG_3085|M.smegmatis_MC2_155 VAADRIPDGKMGLDIGPGSVERFTALLSNAKTVFWNGPMGVFEFPAFAAG
TH_0427|M.thermoresistible__bu VAADQIPDGKMGLDIGPGTVQRFTNLLSNAKTVFWNGPMGVFEFPAFAAG
MAB_2778c|M.abscessus_ATCC_199 VAADAIPDSKMGLDIGPESVKRFAAVLSNAKTIFWNGPMGVFEFPAFAAG
Mb1472|M.bovis_AF2122/97 VDVGAVPNGLMGLDIGPGSIKRFSTLLSNAGTIFWNGPMGVFEFPAYAAG
Rv1437|M.tuberculosis_H37Rv VDVGAVPNGLMGLDIGPGSIKRFSTLLSNAGTIFWNGPMGVFEFPAYAAG
MLBr_00571|M.leprae_Br4923 VAADAIPDGLMGLDIGPESVKRFATLLSNASTIFWNGPMGVFEFPAYAAG
MMAR_2240|M.marinum_M VAADAIPADTMGLDIGPGSVKRFAALLSNAKTIFWNGPMGVFEFPAYATG
MUL_1829|M.ulcerans_Agy99 VAADAIPADTMGLDIGPGSVKRFAALLSNAKTIFWNGPMGVFEFPAYATG
MAV_3340|M.avium_104 VPADAIPDDLMGLDIGPGSVKRFTALLSNAETIFWNGPMGVFEFPAFAAG
* .. :* . ******* ::.**: :**** *:*************:*:*
Mvan_2704|M.vanbaalenii_PYR-1 TKGVAEAIIGATGKGAFSVVGGGDSAAAVRQLGLPEDGFSHISTGGGASL
Mflv_3709|M.gilvum_PYR-GCK TKGVAEAIIGATGKGAFSVVGGGDSAAAVRQLGLPEDGFSHISTGGGASL
MSMEG_3085|M.smegmatis_MC2_155 TKGVAEAIIGATGKGAFSVVGGGDSAAAVRRLGLPEDGFSHISTGGGASL
TH_0427|M.thermoresistible__bu TKGVAEAIIGATAKGAFSVVGGGDSAAAVRQLGLPEDGFSHISTGGGASL
MAB_2778c|M.abscessus_ATCC_199 TRGVAEAIIAATEKGAFSVVGGGDSAAAVRALDLPDDGFSHISTGGGASL
Mb1472|M.bovis_AF2122/97 TRGVAEAIVAATGKGAFSVVGGGDSAAAVRAMNIPEGAFSHISTGGGASL
Rv1437|M.tuberculosis_H37Rv TRGVAEAIVAATGKGAFSVVGGGDSAAAVRAMNIPEGAFSHISTGGGASL
MLBr_00571|M.leprae_Br4923 TRGVAEAIVAVTGKGAFSVVGGGDSAAAMRALSLPEGSVSHLSTGGGASL
MMAR_2240|M.marinum_M TRGIAEAIVAATGKGAFSVVGGGDSAAAVRALGISEGSFSHISTGGGASL
MUL_1829|M.ulcerans_Agy99 TRGIAEAIVAATGKGAFSVVGGGDSAAAVRALGISEGSFSHISTGGGASL
MAV_3340|M.avium_104 TKGLAEAIAAATGKGAFSVVGGGDSAAAVRALGIPESGFSHISTGGGASL
*:*:**** ..* ***************:* :.:.:...**:********
Mvan_2704|M.vanbaalenii_PYR-1 EYLEGKELPGIQVLE-------------
Mflv_3709|M.gilvum_PYR-GCK EYLEGKELPGIEVLN-------------
MSMEG_3085|M.smegmatis_MC2_155 EYLEGKELPGIQVLES------------
TH_0427|M.thermoresistible__bu EYLEGKVLPGLEVLES------------
MAB_2778c|M.abscessus_ATCC_199 EYLEGKVLPGIDVLED------------
Mb1472|M.bovis_AF2122/97 EYLEGKTLPGIEVLSREQPT----GGVL
Rv1437|M.tuberculosis_H37Rv EYLEGKTLPGIEVLSREQPT----GGVL
MLBr_00571|M.leprae_Br4923 EYLEGKTLPGIEVLGREQPR----GGDS
MMAR_2240|M.marinum_M EYLEGKTLPGIEVLGRPQPGQTEGGGPA
MUL_1829|M.ulcerans_Agy99 EYLEGKTLPGIEVLGRPQPGQTEGGGPA
MAV_3340|M.avium_104 EYLEGKALPGIEVLGRPQPT----GGAA
****** ***::**