For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPRAVLVGLPGSGKSTIGRRLAKTLQLPLLDTDVAIEKATGRTIADIFATDGEAEFRRIEESVIRDALA THDGVLSLGGGAVTTEGVREALCGHTVIYLEISAAEGVRRTGGSTVRPLLAGPDRNEKFKALMAERVPLY RQVATMRINTNHRNPGAVVRHIVARLENPEVKPERRRRRPVWRRAPLSLTASPSTEAPPSPAAVAARKST GSPCARKDMQ
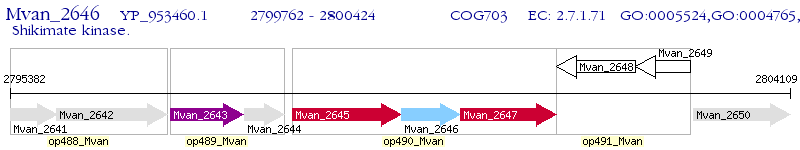
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2646 | aroK | - | 100% (220) | shikimate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2568c | aroK | 2e-74 | 81.07% (169) | shikimate kinase |
| M. gilvum PYR-GCK | Mflv_3757 | aroK | 1e-108 | 88.18% (220) | shikimate kinase |
| M. tuberculosis H37Rv | Rv2539c | aroK | 2e-74 | 81.07% (169) | shikimate kinase |
| M. leprae Br4923 | MLBr_00517 | aroK | 2e-64 | 73.65% (167) | shikimate kinase |
| M. abscessus ATCC 19977 | MAB_2842c | aroK | 1e-71 | 70.50% (200) | shikimate kinase |
| M. marinum M | MMAR_2176 | aroK | 1e-71 | 77.51% (169) | shikimate kinase AroK |
| M. avium 104 | MAV_3416 | aroK | 3e-70 | 77.71% (166) | shikimate kinase |
| M. smegmatis MC2 155 | MSMEG_3031 | aroK | 2e-81 | 69.20% (237) | shikimate kinase |
| M. thermoresistible (build 8) | TH_0109 | aroK | 2e-85 | 77.88% (217) | SHIKIMATE KINASE AROK (SK) |
| M. ulcerans Agy99 | MUL_1760 | aroK | 2e-71 | 76.92% (169) | shikimate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2646|M.vanbaalenii_PYR-1 MAPRAVLVGLPGSGKSTIGRRLAKTLQLPLLDTDVAIEKATGRTIADIFA
Mflv_3757|M.gilvum_PYR-GCK MAPKAVLIGLPGSGKSTIGRRLAKSLGVPLLDTDNAIEETTGRTIADIFA
TH_0109|M.thermoresistible__bu MAPRAVLVGLPGSGKSTIGRRLAKALDLTLLDTDVVIEERTGRTIADIFA
MSMEG_3031|M.smegmatis_MC2_155 MAPRAVLIGLPGSGKSTIGRRLAKVLDVSMLDTDAVIEETAGRTIADIFA
MMAR_2176|M.marinum_M MAPKAVLVGLPGSGKSTIGRRLAKALGVGLLDTDAAIEQQAGRSIAEIFA
MUL_1760|M.ulcerans_Agy99 MAPKAVLVGLPGSGKSTIGRRLAKALGVSLLDTDAAIEQQAGRSIAEIFA
MLBr_00517|M.leprae_Br4923 MAPKAVLVGLPGAGKSTIGRRLSKALGVSLLDTDAAIEKQTGRSIADIFA
Mb2568c|M.bovis_AF2122/97 MAPKAVLVGLPGSGKSTIGRRLAKALGVGLLDTDVAIEQRTGRSIADIFA
Rv2539c|M.tuberculosis_H37Rv MAPKAVLVGLPGSGKSTIGRRLAKALGVGLLDTDVAIEQRTGRSIADIFA
MAV_3416|M.avium_104 MAPKAVLIGLPGSGKSTIGRRLAKALGVGFLDTDAAIEQRTGRPIAEIFA
MAB_2842c|M.abscessus_ATCC_199 MSPKAVLVGLPGSGKSTIGRRLAKALGVNVYDTDTGIETEAGRTIAQIFA
*:*:***:****:*********:* * : . *** ** :**.**:***
Mvan_2646|M.vanbaalenii_PYR-1 TDGEAEFRRIEESVIRDALATHDGVLSLGGGAVTTEGVREALCGHTVIYL
Mflv_3757|M.gilvum_PYR-GCK TDGESEFRRIEEQVIRDALATHDGVLSLGGGAVTTEGVREALAGHTVIYL
TH_0109|M.thermoresistible__bu TDGEPEFRRIEEEVIREALETHDGVLSLGGGAVTTPGVREALVGHTVIYL
MSMEG_3031|M.smegmatis_MC2_155 TDGEKEFRRIEEEVIRSALQTHDGVLSLGGGAVTTPGVREALAGHTVVYL
MMAR_2176|M.marinum_M TDGEEEFRRIEEEVVRAALADHDGVLSLGGGAVTSPGVRSALDGHTVVYL
MUL_1760|M.ulcerans_Agy99 TDGEEEFRRIEEEVVRAALVDHDGVLSLGGGAVTSPGVRSALDGHTVVYL
MLBr_00517|M.leprae_Br4923 IDGEEEFRRIEEGVVRAALVEHDGVVSLGGGAVTSPGVCAALAGHIVIYL
Mb2568c|M.bovis_AF2122/97 TDGEQEFRRIEEDVVRAALADHDGVLSLGGGAVTSPGVRAALAGHTVVYL
Rv2539c|M.tuberculosis_H37Rv TDGEQEFRRIEEDVVRAALADHDGVLSLGGGAVTSPGVRAALAGHTVVYL
MAV_3416|M.avium_104 TDGEREFRRIEEEVVRAALTEHDGVLSLGGGAVTSPGVREALAGHTVVYL
MAB_2842c|M.abscessus_ATCC_199 NDGEPEFRRIEESVIRQALDQQDGVVSLGGGAVLTPGVREALAGHTVVYL
*** ******* *:* ** :***:******* : ** ** ** *:**
Mvan_2646|M.vanbaalenii_PYR-1 EISAAEGVRRTGGSTVRPLLAGPDRNEKFKALMAERVPLYRQVATMRINT
Mflv_3757|M.gilvum_PYR-GCK EISAAEGVRRTGGSTVRPLLAGPDRGEKFKALMAERVPLYRQVATMRVNT
TH_0109|M.thermoresistible__bu EIGLAEGVRRTGGSTVRPLLAGGDRTEKFRKLMAERGPLYRQVATMRINT
MSMEG_3031|M.smegmatis_MC2_155 EISAAEGVRRTGGSTVRPLLAGGDRAEKYRKLMAERVPLYRKVATIRINT
MMAR_2176|M.marinum_M EISAAEGVRRTGGSNVRPLLAGPDRAEKFRALMSQRIPLYRRVSTIRVDT
MUL_1760|M.ulcerans_Agy99 EISAAEGVRRTGGSNVRPLLAGPDRAEKFRALMSQRIPLYRRVSTIRVDT
MLBr_00517|M.leprae_Br4923 EINAEEAMRRACGSTVRPLLAGPDRAEKFQDLMARRVPLYRRVATIRVDT
Mb2568c|M.bovis_AF2122/97 EISAAEGVRRTGGNTVRPLLAGPDRAEKYRALMAKRAPLYRRVATMRVDT
Rv2539c|M.tuberculosis_H37Rv EISAAEGVRRTGGNTVRPLLAGPDRAEKYRALMAKRAPLYRRVATMRVDT
MAV_3416|M.avium_104 EISATEGVRRTGGNTVRPLLAGPDRAEKYRALLAERSPLYRRAATIRVDT
MAB_2842c|M.abscessus_ATCC_199 EISAAEGIRRTGGSVVRPLLAGPDRAEKYHALMSQRVPLYREVATIKVNT
**. *.:**: *. ******* ** **:: *::.* ****..:*::::*
Mvan_2646|M.vanbaalenii_PYR-1 NHRNPGAVVRHIVARLEN--PE----VKPER-------RRRRP----VWR
Mflv_3757|M.gilvum_PYR-GCK NHRNPGAVVRYICARLEN--PD----AEPDR-------RRRRS----VWR
TH_0109|M.thermoresistible__bu NRRNPGAVVRQIVSRLEDSRPENAQNTSPRR-------RRRRRGGSGRSR
MSMEG_3031|M.smegmatis_MC2_155 NRRNPGAVVRTIVARMEN--PQQAKPVAAARTPKPGKPRRRRR---PPWR
MMAR_2176|M.marinum_M NRRNPGAVVRYIMSRLDDPTPN----------------------------
MUL_1760|M.ulcerans_Agy99 NRRNPGAVVRYIMSRLDDPTPN----------------------------
MLBr_00517|M.leprae_Br4923 NCHNLGAVVRYIMARLQAQLATPVS-------------------------
Mb2568c|M.bovis_AF2122/97 NRRNPGAVVRHILSRLQVPSPS----------------------------
Rv2539c|M.tuberculosis_H37Rv NRRNPGAVVRHILSRLQVPSPS----------------------------
MAV_3416|M.avium_104 NRRNPGAVVRYIVSRLPATDAC----------------------------
MAB_2842c|M.abscessus_ATCC_199 DRRNPGAVVRMIVSRLENPEGS----------------------------
: :* ***** * :*:
Mvan_2646|M.vanbaalenii_PYR-1 RA----------PLSLTASPS--------TEAPPSPAAVAARKSTGSPCA
Mflv_3757|M.gilvum_PYR-GCK RA----------PLSLTASPS--------TEAPPSPATVAARKARGSSCM
TH_0109|M.thermoresistible__bu RE----------PATLTAEPS--------TDNPPTPAALAVRHAE-----
MSMEG_3031|M.smegmatis_MC2_155 RTGKNGKNGSAEPKAETAGDSGIASAATTTKVTPTPAALAARNVER----
MMAR_2176|M.marinum_M -------------TSPSSTAS---------GAAT----------------
MUL_1760|M.ulcerans_Agy99 -------------TSPSSTAS---------GAAT----------------
MLBr_00517|M.leprae_Br4923 ---------GGDRKSSEAERS---------GAPLRKSSEVVK--------
Mb2568c|M.bovis_AF2122/97 ----------------EAAT------------------------------
Rv2539c|M.tuberculosis_H37Rv ----------------EAAT------------------------------
MAV_3416|M.avium_104 ----------------RAAT------------------------------
MAB_2842c|M.abscessus_ATCC_199 ------------AGSASSRRRRPRRRPRSRRRSAGAAAVTQTNTSGTETR
:
Mvan_2646|M.vanbaalenii_PYR-1 RKDMQ-
Mflv_3757|M.gilvum_PYR-GCK RKDPQ-
TH_0109|M.thermoresistible__bu RRE---
MSMEG_3031|M.smegmatis_MC2_155 HND---
MMAR_2176|M.marinum_M ------
MUL_1760|M.ulcerans_Agy99 ------
MLBr_00517|M.leprae_Br4923 ------
Mb2568c|M.bovis_AF2122/97 ------
Rv2539c|M.tuberculosis_H37Rv ------
MAV_3416|M.avium_104 ------
MAB_2842c|M.abscessus_ATCC_199 ERADND