For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTNWLDEVFGVAKPVIAMLHLSALPGDPGYDSAGGIAAVVNRARTELEALQGGGVDGVMISNEFSLPYLT KTEPITAITMARIIGELLPDISVPFGVNVLWDGRASIDLAVATGAKWVREIFTGVYASDFGLWDTNVGEV ARHRARVGGADVKLLFNIVPESAQYLADRDLASITRTTVFATLPDAICVSGATAGAPTDTEALRIVKAAA GEVPVFVNTGVRAENVAAQLSVADGAVVGTYFKKDGLFANAVEKSRVEALMGAAKEFRAQR*
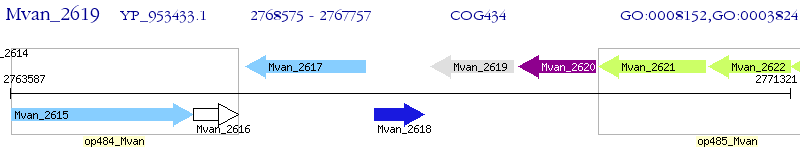
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2619 | - | - | 100% (272) | photosystem I assembly BtpA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3780 | - | 1e-139 | 91.48% (270) | photosystem I assembly BtpA |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3114 | - | 1e-122 | 81.65% (267) | hypothetical protein MSMEG_3114 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2619|M.vanbaalenii_PYR-1 MTTNWLDEVFGVAKPVIAMLHLSALPGDPGYDSAGGIAAVVNRARTELEA
Mflv_3780|M.gilvum_PYR-GCK MTTTWLDEVFNVTKPVIAMLHLSALPGDPGYDSAGGIAAVVNRARTELEA
MSMEG_3114|M.smegmatis_MC2_155 -MTSWLDDVFAVRKPVIAMLHLSALPGDPGFDSAAGIGAVVERAKSELAA
*.***:** * *****************:***.**.***:**::** *
Mvan_2619|M.vanbaalenii_PYR-1 LQGGGVDGVMISNEFSLPYLTKTEPITAITMARIIGELLPDISVPFGVNV
Mflv_3780|M.gilvum_PYR-GCK LQSGGVDAVMISNEFSLPYLTKTEPITAITMARIIGELLPDISVPYGVNV
MSMEG_3114|M.smegmatis_MC2_155 LQEGGVDGVMISNEFSLPYLTETEPITAITMARIIGELLPEISVPYGVNV
** ****.*************:******************:****:****
Mvan_2619|M.vanbaalenii_PYR-1 LWDGRASIDLAVATGAKWVREIFTGVYASDFGLWDTNVGEVARHRARVGG
Mflv_3780|M.gilvum_PYR-GCK LWDARASIDLAVATGAKFVREIFTGVYASDFGLWDTNVGEVARHRARVGG
MSMEG_3114|M.smegmatis_MC2_155 LWDGRASIDLAVATGAQWVREIFTGVYASDFGLWNTNVGAVARHRHRIGG
***.************::****************:**** ***** *:**
Mvan_2619|M.vanbaalenii_PYR-1 ADVKLLFNIVPESAQYLADRDLASITRTTVFATLPDAICVSGATAGAPTD
Mflv_3780|M.gilvum_PYR-GCK AGVKLLYNIVPESATYLADRDLASITRTTVFATAPDGICVSGATAGSPTD
MSMEG_3114|M.smegmatis_MC2_155 DHVKLLFNIVPESAVYLADRDLASITATTVFATKPDAICVSGLTAGASTD
****:******* *********** ****** **.***** ***:.**
Mvan_2619|M.vanbaalenii_PYR-1 TEALRIVKAAAGEVPVFVNTGVRAENVAAQLSVADGAVVGTYFKKDGLFA
Mflv_3780|M.gilvum_PYR-GCK TDALRVVKSAAGDVPVFVNTGVRAENVADQLSVADGAVVGTYFKKDGLFA
MSMEG_3114|M.smegmatis_MC2_155 RQALQVVKNAAGEVPVFVNTGVRAHNAAEQLAIADGAVVGTYFKEDGVFE
:**::** ***:***********.*.* **::***********:**:*
Mvan_2619|M.vanbaalenii_PYR-1 NAVEKSRVEALMGAAKEFRAQR-
Mflv_3780|M.gilvum_PYR-GCK NAAEKSRVEELMGAARDFRATLG
MSMEG_3114|M.smegmatis_MC2_155 NRAVVSRVSELMDAVKGFRKTL-
* . ***. **.*.: **