For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GTGQQPGEHGCRLRSPPPETNHHDLLCMVMHPCERVGLDFVDTAPYRFVSTVDLAITPEQLFEVLADETS WPHWASVITKVTWTSPEPRGVGTTRTVDMRGGITGAEEFLAWEPFTRMAFRFNESSTGSISAFAEDYHVV PTAGGCHLTWIMALKPNGPAGRFGLAAGKPVMGWLFQRFLHNLRRYTDERFATR*
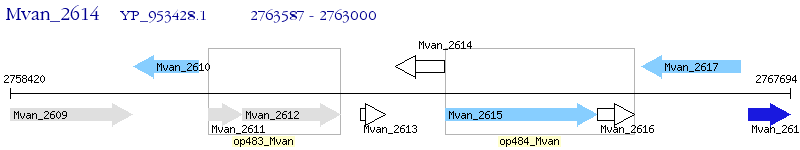
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2614 | - | - | 100% (195) | hypothetical protein Mvan_2614 |
| M. vanbaalenii PYR-1 | Mvan_2251 | - | 3e-18 | 37.60% (125) | hypothetical protein Mvan_2251 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2604 | - | 7e-62 | 68.71% (163) | hypothetical protein Mb2604 |
| M. gilvum PYR-GCK | Mflv_3786 | - | 8e-79 | 83.33% (162) | hypothetical protein Mflv_3786 |
| M. tuberculosis H37Rv | Rv2574 | - | 7e-62 | 68.71% (163) | hypothetical protein Rv2574 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3139c | - | 4e-12 | 39.33% (89) | hypothetical protein MAB_3139c |
| M. marinum M | MMAR_2156 | - | 2e-55 | 63.25% (166) | hypothetical protein MMAR_2156 |
| M. avium 104 | MAV_3449 | - | 7e-61 | 67.88% (165) | hypothetical protein MAV_3449 |
| M. smegmatis MC2 155 | MSMEG_3001 | - | 7e-72 | 73.91% (161) | hypothetical protein MSMEG_3001 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1741 | - | 5e-55 | 62.05% (166) | hypothetical protein MUL_1741 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2614|M.vanbaalenii_PYR-1 MGTGQQPGEHGCRLRSPPPETNHHDLLCMVMHPCERVGLDFVDT----AP
Mflv_3786|M.gilvum_PYR-GCK ------------------------------MHSCERVGLDFIDS----AP
MSMEG_3001|M.smegmatis_MC2_155 ---------------------------MARTYECERVDLDFIDR----AP
Mb2604|M.bovis_AF2122/97 ------------------------------MYPCERVGLSFTET----AP
Rv2574|M.tuberculosis_H37Rv ------------------------------MYPCERVGLSFTET----AP
MAV_3449|M.avium_104 ------------------------------MFDCERVGLDFIDS----AP
MMAR_2156|M.marinum_M ------------------------------MFPCERVDLSFVDPDSGQAP
MUL_1741|M.ulcerans_Agy99 ------------------------------MFPCERVDLSFVDPDSGQAP
MAB_3139c|M.abscessus_ATCC_199 -----------------------------MVMPSFKLEP-LVESDFNTSD
. :: : : :
Mvan_2614|M.vanbaalenii_PYR-1 YRFVSTVDLAITPEQLFEVLADETSWPHWASVITKVTWTSPEPRGVGTTR
Mflv_3786|M.gilvum_PYR-GCK YRFVSTVDLAITPEQLFEVLADETSWPHWATVITDVTWTTPEPRGVGTTR
MSMEG_3001|M.smegmatis_MC2_155 FRFVSTVDLPITGEQLFEVLADETSWPKWATVITDVEWTSPQPRGVGTTR
Mb2604|M.bovis_AF2122/97 YLFRNTVDLAITPEQLFEVLADPQAWPRWATVITKVTWTSPEPFGAGTTR
Rv2574|M.tuberculosis_H37Rv YLFRNTVDLAITPEQLFEVLADPQAWPRWATVITKVTWTSPEPFGAGTTR
MAV_3449|M.avium_104 VVFRNSVDLAITPEQLFEVLSDAAAWPRWATVITKVTWTSPEPRGVGTTR
MMAR_2156|M.marinum_M FLYRNSVDLAITPEQLFEVLSDAESWPSWAPVITKVTWTSPPPFRVGTTR
MUL_1741|M.ulcerans_Agy99 FLYRNSVDLAITPEQLFEVLSDAESWPSWAPVITKVTWTSPPPFRIGTTR
MAB_3139c|M.abscessus_ATCC_199 FVYTFSTPLPKDAATVWAELNGESPL-HWCKAINQIGWTSPEPRGVGSTR
: :. *. :: * . . *. .*..: **:* * *:**
Mvan_2614|M.vanbaalenii_PYR-1 TVDMRG-GITGAEEFLAWEPFT---RMAFRFNESSTGSISAFAEDYHVVP
Mflv_3786|M.gilvum_PYR-GCK TVQMRG-GITGDEVFLAWEPFS---RMAFRFNAASTRSISAFAEDYRVVP
MSMEG_3001|M.smegmatis_MC2_155 TVHMRG-NIVGHEEFLAWEPFS---RMAFRFNTTTSGAISAFAEDYRVVE
Mb2604|M.bovis_AF2122/97 IVEMRG-GIVGDEEFISWEPFT---RMAFRFNECSTRAVGAFAEDYRVQA
Rv2574|M.tuberculosis_H37Rv IVEMRG-GIVGDEEFISWEPFT---RMAFRFNECSTRAVGAFAEDYRVQA
MAV_3449|M.avium_104 VVEMRG-GIVGNEEFLAWEPFT---HMAFRFNECSTRSVAAFAEDYRVQV
MMAR_2156|M.marinum_M TVEMRG-GLVGNEEFIAWEPFS---HMAFRFNECSTQAVAAFAEDYRVEV
MUL_1741|M.ulcerans_Agy99 TVEMRG-GLVGNEEFIAWEPFS---HMAFRFNECSTQAVAAFAEDYRVEA
MAB_3139c|M.abscessus_ATCC_199 TAKLALPGAAVNERFIVWEESPERYRNAFTVETATFPGTKRFGELYEVVG
..: . . * *: ** . : ** .: : . *.* *.*
Mvan_2614|M.vanbaalenii_PYR-1 TAGGCHLTWIMALKPNGPAGRFGLAAGKPVMGWLFQRFLHNLRRYTDERF
Mflv_3786|M.gilvum_PYR-GCK TQNGCHLTWIMALQPNGVAGRAGLFLGRPVMARMFQKFLHNLRDYTDERF
MSMEG_3001|M.smegmatis_MC2_155 TPQGCHLTWVMAIRPSGPAGRLGMLLGRPLMAWMFQRFLHNLRRYSDARY
Mb2604|M.bovis_AF2122/97 IPGGCRLTWTMAQKLAGPA-RPALFVFRPLLNLALRRFLRNLRRYTDARF
Rv2574|M.tuberculosis_H37Rv IPGGCRLTWTMAQKLAGPA-RPALFVFRPLLNLALRRFLRNLRRYTDARF
MAV_3449|M.avium_104 IPGGCRLTWTMAQKPAGPA-KLAMAVVGPLLNLALRRFLRNLRSYTDS--
MMAR_2156|M.marinum_M IPGGCRLTWTMAQKPAGPA-RLGMMLFGPLLNLGLRRFLRKLRSYTDARF
MUL_1741|M.ulcerans_Agy99 IPGGCRLTWTMAQKPAGPA-RLGMMLFGPLLNLGLRHFLRKLRSYTDARF
MAB_3139c|M.abscessus_ATCC_199 TATGSLFTWSFFIEP--------SLGFTRHLKPVIRRGLSGLITDTQKHF
*. :** : . : ::: * * ::
Mvan_2614|M.vanbaalenii_PYR-1 ATR---
Mflv_3786|M.gilvum_PYR-GCK GG----
MSMEG_3001|M.smegmatis_MC2_155 PSARSA
Mb2604|M.bovis_AF2122/97 AAAQQS
Rv2574|M.tuberculosis_H37Rv AAAQQS
MAV_3449|M.avium_104 LATRQP
MMAR_2156|M.marinum_M EVPQQR
MUL_1741|M.ulcerans_Agy99 EVPQQR
MAB_3139c|M.abscessus_ATCC_199 A-----