For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLVVFLPLLIIMGAFMYFASRRQKKAMQATIDLHNSLQIGDRVHTTSGLRATITGITDDYVDLEIAPGV VTTWMKLAVRDRIVDDVEDDDDGTLDEAESSAVELNKSPNEKTDS
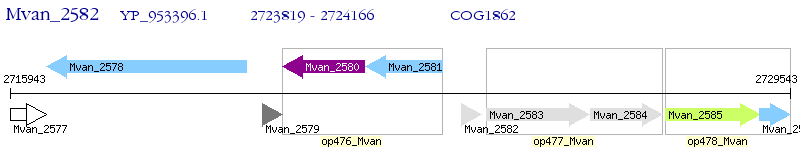
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2582 | yajC | - | 100% (115) | preprotein translocase subunit YajC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2619c | yajC | 5e-35 | 74.47% (94) | preprotein translocase subunit YajC |
| M. gilvum PYR-GCK | Mflv_3817 | yajC | 9e-47 | 81.58% (114) | preprotein translocase subunit YajC |
| M. tuberculosis H37Rv | Rv2588c | yajC | 5e-35 | 74.47% (94) | preprotein translocase subunit YajC |
| M. leprae Br4923 | MLBr_00486 | yajC | 1e-31 | 67.35% (98) | preprotein translocase subunit YajC |
| M. abscessus ATCC 19977 | MAB_2881c | yajC | 4e-40 | 70.37% (108) | preprotein translocase subunit YajC |
| M. marinum M | MMAR_2119 | yajC | 1e-38 | 70.64% (109) | membrane protein secretion factor YajC |
| M. avium 104 | MAV_3469 | yajC | 8e-37 | 78.41% (88) | preprotein translocase subunit YajC |
| M. smegmatis MC2 155 | MSMEG_2960 | yajC | 1e-41 | 84.78% (92) | preprotein translocase subunit YajC |
| M. thermoresistible (build 8) | TH_1439 | - | 6e-38 | 77.66% (94) | PROBABLE CONSERVED MEMBRANE PROTEIN SECRETION FACTOR YAJC |
| M. ulcerans Agy99 | MUL_1724 | yajC | 9e-39 | 70.64% (109) | preprotein translocase subunit YajC |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2119|M.marinum_M -----------------------------------MESFVLFLPFLLIMG
MUL_1724|M.ulcerans_Agy99 -----------------------------------MESFVLFLPFLLIMG
Mb2619c|M.bovis_AF2122/97 -----------------------------------MESFVLFLPFLLIMG
Rv2588c|M.tuberculosis_H37Rv -----------------------------------MESFVLFLPFLLIMG
MAV_3469|M.avium_104 ---------------------------------MSMESLILFLPFLLIMG
MLBr_00486|M.leprae_Br4923 -----------------------------------MESLVLLLLFLLIMG
Mvan_2582|M.vanbaalenii_PYR-1 ------------------------------------MDLVVFLPLLIIMG
Mflv_3817|M.gilvum_PYR-GCK ------------------------------------MDLVVFLPLLIIMG
MSMEG_2960|M.smegmatis_MC2_155 ------------------------------------MDLVIFLPLLIIMG
TH_1439|M.thermoresistible__bu VQSAPQCRNGTLVTYEDSCARSGARDHKTDTKDKRFMDLVVFLPLLIVLG
MAB_2881c|M.abscessus_ATCC_199 -----------------------------------MESIIVFLPLILVMG
.::::* :::::*
MMAR_2119|M.marinum_M GFMYFASRRQRKAMQATIDLHESLEPGDRVHTTSGLQATVVGITEDTVDL
MUL_1724|M.ulcerans_Agy99 GFMYFASRRQRKAMQATIDLHESLEPGDRVHTTSGLQATVVGITEDTVDL
Mb2619c|M.bovis_AF2122/97 GFMYFASRRQRRAMQATIDLHDSLQPGERVHTTSGLEATIVAIADDTIDL
Rv2588c|M.tuberculosis_H37Rv GFMYFASRRQRRAMQATIDLHDSLQPGERVHTTSGLEATIVAIADDTIDL
MAV_3469|M.avium_104 GFMYFASRRQRRAMQATIELHESLRPGDRVHTTSGLQATVVGLTDDTVDL
MLBr_00486|M.leprae_Br4923 GFMFFASRRQRRSMQATIDLYNSLQPGDRVNTTSGLQATIIVVGDDTVDL
Mvan_2582|M.vanbaalenii_PYR-1 AFMYFASRRQKKAMQATIDLHNSLQIGDRVHTTSGLRATITGITDDYVDL
Mflv_3817|M.gilvum_PYR-GCK AFMYFASRRQKKAMQATIDLHNSLEIGDRIHTTSGLQGTITGIGDDYVDV
MSMEG_2960|M.smegmatis_MC2_155 AFMFFASRRQKKAMQATIDLHNSLQIGDQIHTTSGLKGTITGISDDYVDL
TH_1439|M.thermoresistible__bu AFMFFASRRQKKAMQAMIDLHESLRVGDQVHTTAGLKGTIRAIDDDYVDL
MAB_2881c|M.abscessus_ATCC_199 AFLFFANRRQRKALDATIELHASLAIGDRVHTTSGLQGTITAVTDDTVDL
.*::**.***:::::* *:*: ** *::::**:**..*: : :* :*:
MMAR_2119|M.marinum_M EIAPGVVTTWMKLAVRDRILPDDDSDGDLDAE---SPEISEISESHTAKE
MUL_1724|M.ulcerans_Agy99 EIAPGVVTTWMKLAVRDRILPDDDSDGDLDAE---SPEISEISESHTAKE
Mb2619c|M.bovis_AF2122/97 EIAPGVVTTWMKLAIRDRILPDDDIDEELNEDLDKDVDDV-AGERRVTND
Rv2588c|M.tuberculosis_H37Rv EIAPGVVTTWMKLAIRDRILPDDDIDEELNEDLDKDVDDV-AGERRVTND
MAV_3469|M.avium_104 EIAPGVVTTWMKLAIRDRIVPDD--EDDL-----VDGDDS-AGEGSVSKD
MLBr_00486|M.leprae_Br4923 EIAPGVVTTWMKLAIRDRILPDDAYMDEHEAE---PGDFVYCDELEESDG
Mvan_2582|M.vanbaalenii_PYR-1 EIAPGVVTTWMKLAVRDRIVDDVEDDDDGTLDEAESSAVELNKSPNEKTD
Mflv_3817|M.gilvum_PYR-GCK EIAPGVVTTWMKLAVRDRIVDDVED--DAELDD--ESAVEITERPNLNKD
MSMEG_2960|M.smegmatis_MC2_155 EIAPGVITTWMKLAVRDRIDSGLAQ-----DDD--ELSEHTNDTDGYKND
TH_1439|M.thermoresistible__bu EIAPGVVTTWMKLAVRDRIEPQTAD-----TDD--VTDVSTSDSTGQDRL
MAB_2881c|M.abscessus_ATCC_199 EIAPGVVTRWMKLAVRDKIVDEVEDS--AAADDGTVRDVESSGVELTKE-
******:* *****:**:*
MMAR_2119|M.marinum_M L-
MUL_1724|M.ulcerans_Agy99 L-
Mb2619c|M.bovis_AF2122/97 S-
Rv2588c|M.tuberculosis_H37Rv S-
MAV_3469|M.avium_104 S-
MLBr_00486|M.leprae_Br4923 SS
Mvan_2582|M.vanbaalenii_PYR-1 S-
Mflv_3817|M.gilvum_PYR-GCK --
MSMEG_2960|M.smegmatis_MC2_155 --
TH_1439|M.thermoresistible__bu S-
MAB_2881c|M.abscessus_ATCC_199 --