For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDETGRPGITRRWARWRDRLRERRAADLVYRIAVGVVGLLVLAVGILAIPYPGPGWAIVFVGLAILATE FDWARRLLAYARARYDTAMAWFKRQGLWVQALGALFTFAIVMGTLWLLGALAFVGELVGIEYRWLKSPIG IGE
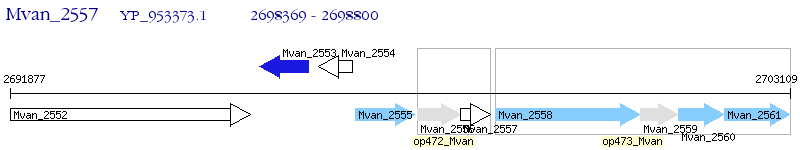
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2557 | - | - | 100% (143) | hypothetical protein Mvan_2557 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1959c | - | 3e-06 | 28.33% (120) | hypothetical protein Mb1959c |
| M. gilvum PYR-GCK | Mflv_3839 | - | 1e-61 | 77.70% (139) | hypothetical protein Mflv_3839 |
| M. tuberculosis H37Rv | Rv1924c | - | 3e-06 | 28.33% (120) | hypothetical protein Rv1924c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2834 | - | 6e-07 | 29.57% (115) | hypothetical protein MMAR_2834 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2930 | - | 2e-42 | 63.08% (130) | hypothetical protein MSMEG_2930 |
| M. thermoresistible (build 8) | TH_4657 | - | 1e-39 | 56.82% (132) | PUTATIVE putative conserved hypothetical protein TIGR02611 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2557|M.vanbaalenii_PYR-1 ----------------------MSDETGRPGITRRWAR-WRDRLRERRAA
Mflv_3839|M.gilvum_PYR-GCK ----------------------MTDTD-KPGFTRRWAR-RRDRLRERPVV
MSMEG_2930|M.smegmatis_MC2_155 ----------------------MNVKEKLSEYRDQWVH-QRERLRKRPLA
TH_4657|M.thermoresistible__bu VSVQPSEQHRPAPETDGFSAAPTSGWRHRLTARRQWLAARREQLRRRRIL
Mb1959c|M.bovis_AF2122/97 ---------------------MDPADVINPTSTRDAALARVLAYRQRVRA
Rv1924c|M.tuberculosis_H37Rv ---------------------MDPADVINPTSTRDAALARVLAYRQRVRA
MMAR_2834|M.marinum_M -------MPSGPVDRLEAGVSVDSGDCNPAATTRDAAFARVLAYRDRARA
* *
Mvan_2557|M.vanbaalenii_PYR-1 D-LVYRIAVGVVGLLVLAVGI-LAIPYPGPGWAIVFVGLAILATEFDWAR
Mflv_3839|M.gilvum_PYR-GCK D-LTYRISVGVVGLVVLGLGI-LAIPYPGPGWAIVFLGLAILATEFDWAK
MSMEG_2930|M.smegmatis_MC2_155 N-FVYRSVIGVVGTVVLAVGV-VAIPYPGPGWAIVFLGLAILATEFRFAE
TH_4657|M.thermoresistible__bu N-AVYRSTVGVVGLVVVVIGI-IAIPYPGPGWAIFFLGLALLATEFAWAQ
Mb1959c|M.bovis_AF2122/97 RPLLIRATLAVVGGGLFVVSLPMIVLLPELGIPALLVAFRLLAVEAQWAV
Rv1924c|M.tuberculosis_H37Rv RPLLIRATLAVVGGGLFVVSLPMIVLLPELGIPALLVAFRLLAVEAQWAV
MMAR_2834|M.marinum_M NPMVLRVLLAVFGAALLIASVPLMVLLPELGIPAVLVAFRVLAIESDWAA
* :.*.* :. .: : : * * . .::.: :** * :*
Mvan_2557|M.vanbaalenii_PYR-1 RLLAYARARYDTAMAWFKRQGLWVQALGALFTFAIVMGTLWLLGALAFVG
Mflv_3839|M.gilvum_PYR-GCK RLLGYARRRYDAVMDWFKQQGLWVQALGALFTCAVVVGTLWLFGALAFVG
MSMEG_2930|M.smegmatis_MC2_155 VALRFIRRRYDEVMAWFARQHIAVKALSAGFTAAVVLATLWGLGALYWAG
TH_4657|M.thermoresistible__bu RSLRFTKARYDAVMDWLGRQHPAVQALGAVLTAVVVIVTLWVVGAVGWGA
Mb1959c|M.bovis_AF2122/97 RAYAWTDWRFTQLREWFHRQVLVTRAAILVGLFLAAVALVWLLVYEF---
Rv1924c|M.tuberculosis_H37Rv RAYAWTDWRFTQLREWFHRQVLVTRAAILVGLFLAAVALVWLLVYEF---
MMAR_2834|M.marinum_M RAYAWTDWRFAQLRDWFHRKSAIVRAAILTALLLVAAALVWVLIVEF---
: *: *: :: .:* . :* .
Mvan_2557|M.vanbaalenii_PYR-1 ELVGIEYRWLKSPIGIGE
Mflv_3839|M.gilvum_PYR-GCK RWFGLEYGWLDSPIGIGS
MSMEG_2930|M.smegmatis_MC2_155 GLVGFEPAWLKSPIGIGS
TH_4657|M.thermoresistible__bu ALFGVDRPWLRSPIGIGS
Mb1959c|M.bovis_AF2122/97 ------------------
Rv1924c|M.tuberculosis_H37Rv ------------------
MMAR_2834|M.marinum_M ------------------