For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDLHFTVVDIAPEPYAVTPILMAHIAITDTDDEPVHAVALRCQVRIEPLRRTYSDDEATGLVDLFGPRER WSTTQHTFLWQHTAAVVPGFTAGTVVQLPLECTYDFEVRAAKYLLALDAGSVPLQFLFSGTIFTQGERGF SVQQVPWDREEHVDMPVSVWRELIRLHYPGTGWLRLTRDTIDALAAYRSARGLLGFDDAISELLAGAREL R*
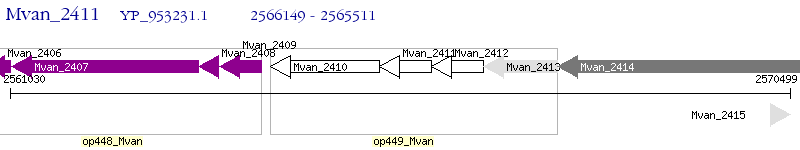
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2411 | - | - | 100% (212) | hypothetical protein Mvan_2411 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1866 | - | 6e-79 | 66.02% (206) | hypothetical protein MMAR_1866 |
| M. avium 104 | MAV_2685 | - | 8e-88 | 70.81% (209) | hypothetical protein MAV_2685 |
| M. smegmatis MC2 155 | MSMEG_2716 | - | 2e-82 | 67.16% (201) | hypothetical protein MSMEG_2716 |
| M. thermoresistible (build 8) | TH_1950 | - | 3e-81 | 66.35% (208) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2716|M.smegmatis_MC2_155 MTSLGADPLAVTFAVLDIAPEPFAVTPILTARLGVSATGEDPVHAIALRC
TH_1950|M.thermoresistible__bu --MIDDRIPDVTFAVLTVHPEPYAITPTLSAQVGIATTGELPVHAIALRA
MMAR_1866|M.marinum_M ---MNEHPLDLTFAVLDVAPEPYAVSPVLTARVGIDTGGEDPVHAIALRC
MAV_2685|M.avium_104 ---MTAPTSDPAFTVVDVTPEPYAVTPVLIARVGVTTSDDEPVHAIALRC
Mvan_2411|M.vanbaalenii_PYR-1 -------MTDLHFTVVDIAPEPYAVTPILMAHIAITDTDDEPVHAVALRC
*:*: : ***:*::* * *::.: .: ****:***.
MSMEG_2716|M.smegmatis_MC2_155 QVRIDPLRRAYTDAEAAGLLDLFGPRDRWNTTQHTFLWQHTAAMVPGFTG
TH_1950|M.thermoresistible__bu QVRIDPVRRGYSDAEAEGLTDLFGSRERWAATQHTFLWQHTATMVPGFTA
MMAR_1866|M.marinum_M QVRIEPLRRPYSDAEADGLVDLFGARERWAITQRTFLWQHCTAMVPGFAG
MAV_2685|M.avium_104 QIRIEPLRRGYSDDEAAGLLDMFGTRERWATTQHTFLWQHCAAMVPGFTG
Mvan_2411|M.vanbaalenii_PYR-1 QVRIEPLRRTYSDDEATGLVDLFGPRERWSTTQHTFLWQHTAAVVPGFTA
*:**:*:** *:* ** ** *:**.*:** **:****** :::****:.
MSMEG_2716|M.smegmatis_MC2_155 ATQIDLALNCTYDFEVAASKYLHALREGTVPLQFLFSGTVFTRGAGGLAV
TH_1950|M.thermoresistible__bu ATRIDLPLVCTYDFDVAASKYLHALRDGAVPLQFLFSGTVFYRGAHGFTV
MMAR_1866|M.marinum_M NTTVALPLDCTYDFEVTAAKYLHALRDGALPLQFLFSGTIFVKSERGFSV
MAV_2685|M.avium_104 STEVSLPLDCTYDFEVTAAKYLHALRDGVVPLQFLFSGTVFRAGARGFSV
Mvan_2411|M.vanbaalenii_PYR-1 GTVVQLPLECTYDFEVRAAKYLLALDAGSVPLQFLFSGTIFTQGERGFSV
* : *.* *****:* *:*** ** * :*********:* . *::*
MSMEG_2716|M.smegmatis_MC2_155 QHIPWDREDHVDMPVAVWQLLMERHFPHTGWLRLDRDTLERLAGYKTQRG
TH_1950|M.thermoresistible__bu RQVPWDREDRYDLPVPVWHDLMRLHYPGTGWLRVQHDTLDALARFRSARG
MMAR_1866|M.marinum_M QQVSWDCEDRYDMPVAVWHQLIAQHYPNSGWVRLGHDTVSALARYKSAHG
MAV_2685|M.avium_104 QQVPWDCEDHHAMPASVWRSLIQQHYPDSGWVRLGHDTVEALAAYKSERG
Mvan_2411|M.vanbaalenii_PYR-1 QQVPWDREEHVDMPVSVWRELIRLHYPGTGWLRLTRDTIDALAAYRSARG
:::.** *:: :*..**: *: *:* :**:*: :**:. ** ::: :*
MSMEG_2716|M.smegmatis_MC2_155 LLSFDEAVGTLL--AREDIP--
TH_1950|M.thermoresistible__bu LLSADDAISALLSAAAEDVR--
MMAR_1866|M.marinum_M LLDLDHAVTALIADAARPEVAQ
MAV_2685|M.avium_104 LLSIDEAVNGLLATAR--EEAR
Mvan_2411|M.vanbaalenii_PYR-1 LLGFDDAISELLAGAR---ELR
**. *.*: *: *