For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRYTVRHTIAGRPAGHSPETPPAEDNSGGWRPLRWLGAPAPGGLALPQAAPSMAWMYSPRGVFVDDKHV VVADSGNHRVLIWHGLPHADEQAADVVLGQPHGYAEGRAAGGRGPERGMNLPTGVAIVDGRLIVADAWHH RLLIWNQVPESDDVAPDVVLGQPDAGSVEPNAGQACSAGTFYWPFGFAILGDRFWVADTGNRRVLGWSGG IPEPGQAADIVLGQPTASAREENRGQAVGPASFRWPHAIAGTSELLLVADAGDHRVLGWSPHPAVDARAN LVLGQPDFVTAQEWPYGPHTGDRFRFPYAIDLDGDCVVVADTANNRILVWDGVQAEALGGRPADHVLAQP DFGSNGENRWSLVGRDTLCWPYGVSLHGDLLAVADSGNNRVMIWRRS
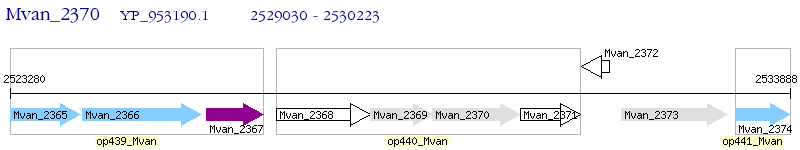
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2370 | - | - | 100% (397) | NHL repeat-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2269 | - | 0.0 | 75.19% (391) | NHL repeat-containing protein |
| M. thermoresistible (build 8) | TH_1026 | - | 1e-175 | 71.07% (401) | NHL repeat protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2370|M.vanbaalenii_PYR-1 MSRYTVRHTIAGRPAGHSPETPPAEDNSGGWRPLRWLGAPAPGGLALPQA
MSMEG_2269|M.smegmatis_MC2_155 -----MRPVADGRPAGRT-AAVLEDELTGGRRPLVWLGAPAPGGLALPPA
TH_1026|M.thermoresistible__bu MTGYTVVRTSLRPPAGHP--APPTADLTGGWTPEVWLGAPAPGGLALPPA
: . ***:. : : :** * ************* *
Mvan_2370|M.vanbaalenii_PYR-1 APSMAWMYSPRGVFVDDKHVVVADSGNHRVLIWHGLPHADEQAADVVLGQ
MSMEG_2269|M.smegmatis_MC2_155 SPTMAWMYSPRGVFVDDEHVVVADSGNHRVLIWHGVPDRDHAPADVVLGQ
TH_1026|M.thermoresistible__bu HPSMAWMYSPRGVYLGDRHLVVADSGNHRVLIWHGLPDRDEQPAHVVLGQ
*:**********::.*.*:***************:*. *. .*.*****
Mvan_2370|M.vanbaalenii_PYR-1 PHGYAEGRAAGGRGPERGMNLPTGVAIVDGRLIVADAWHHRLLIWNQVPE
MSMEG_2269|M.smegmatis_MC2_155 PDGTTEGRAAGGRGPARGMNLPTGVAVIDGRLLVADAWHHRILIWDEVPT
TH_1026|M.thermoresistible__bu PDGVSEGRAAGGRGPQRGMHLPTGVLVHDGRLIVADAWHHRILVWDTVPQ
*.* :********** ***:***** : ****:********:*:*: **
Mvan_2370|M.vanbaalenii_PYR-1 SDDVAPDVVLGQPDAGSVEPNAGQACSAGTFYWPFGFAILGDRFWVADTG
MSMEG_2269|M.smegmatis_MC2_155 RDDVAPDVVLGQPDPESVEPNAGVECSASSFYWPFGFALLGGRFWVADTG
TH_1026|M.thermoresistible__bu RDDVAPDVVLGQPDSSSVAPNQGGGCTASTFYWPYGIAVTGSTFWVADTG
*************. ** ** * *:*.:****:*:*: *. *******
Mvan_2370|M.vanbaalenii_PYR-1 NRRVLGWSGGIPEPGQAADIVLGQPTASAREENRGQAVGPASFRWPHAIA
MSMEG_2269|M.smegmatis_MC2_155 NRRVLAWNNGIPDPGQPADVVLGQPDASAREENRGEPVGPSGFRWPHAIT
TH_1026|M.thermoresistible__bu NRRVLGWTGGIPDPGRPADIVLGQPDATQRAENRGGTAGPASFRWPHALT
*****.*..***:**:.**:***** *: * **** ..**:.******::
Mvan_2370|M.vanbaalenii_PYR-1 GTSELLLVADAGDHRVLGWSPHPAVDARANLVLGQPDFVTAQEWPYGPHT
MSMEG_2269|M.smegmatis_MC2_155 GTGDLLLVADAGDHRILGWSPHPGADVLANVVYGQPDFCSATEWPYGPQT
TH_1026|M.thermoresistible__bu GRADLLLVADAGNHRVLGWSPQPAGDRPAGLLLGQPDFGTCQEWPYGPQS
* .:********:**:*****:*. * *.:: ***** :. ******::
Mvan_2370|M.vanbaalenii_PYR-1 GDRFRFPYAIDLDG----DCVVVADTANNRILVWDGVQAEALGGRPADHV
MSMEG_2269|M.smegmatis_MC2_155 GDRFRFPYGAVVDD----GRLAVADTANNRILLWDSLPDDPVGGRPADHV
TH_1026|M.thermoresistible__bu ADRFRFPYAIALDGTDGTERLAVADTANNRVLLWDGLPAD---GRGADHV
.*******. :*. :.********:*:**.: : ** ****
Mvan_2370|M.vanbaalenii_PYR-1 LAQPDFGSNGENRWSLVGRDTLCWPYGVSLHGDLLAVADSGNNRVMIWRR
MSMEG_2269|M.smegmatis_MC2_155 LAQPDFASNGENRWDLVGRDTLCWPYGLHLHRDLLAVADSGNNRVMLWGR
TH_1026|M.thermoresistible__bu LGQPDFTANGENRWSSVQRDTMCWPYGISLREDRLAVADSGNNRVIVWRR
*.**** :******. * ***:*****: *: * ***********::* *
Mvan_2370|M.vanbaalenii_PYR-1 S
MSMEG_2269|M.smegmatis_MC2_155 R
TH_1026|M.thermoresistible__bu S