For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAVGAGTADLEVLARRVDAAVAALAELDPTARAAAEELKAAVEEVHRAGLTTIVARMRADDAARPLLFD LVDDPVVRMLLTLHGIIRPDPVTLAERALAAVRPQLHSHGGDVTLVRVDDRVAFVRLEGACNGCSMASVT LRELVEAALLQGVPNLEAVEVVSGEATPTLIPVEALRIPPRAEPDGWVNLGSVDSIAVDEIRVLGGDVLV VNIGQKLSAFYNECAHEALPLGDAVFDATAGTLTCPWHGFCYDAESGECLSAPGVQLEQLPLRIDDGDVW VRTGR
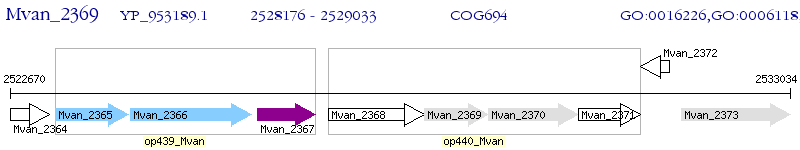
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2369 | - | - | 100% (285) | Rieske (2Fe-2S) domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2413 | - | 3e-20 | 30.37% (270) | NifU domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1868 | - | 6e-15 | 27.46% (284) | hypothetical protein MMAR_1868 |
| M. avium 104 | MAV_2683 | - | 1e-18 | 29.58% (284) | hypothetical protein MAV_2683 |
| M. smegmatis MC2 155 | MSMEG_2268 | - | 1e-102 | 65.96% (285) | hypothetical protein MSMEG_2268 |
| M. thermoresistible (build 8) | TH_1027 | - | 1e-103 | 65.25% (282) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2369|M.vanbaalenii_PYR-1 ---------------MTAVGAGTADLEVLARRVDAAVAALAELDPTARAA
TH_1027|M.thermoresistible__bu MSTTTAAASPAGAAPDAAADAGAREFEALAERVDKAIAALAELDERSRKV
MSMEG_2268|M.smegmatis_MC2_155 ----------------MTAAVEAPELEKLAQRVDNAVRALVDLDPEARKV
MMAR_1868|M.marinum_M ----MADRPETPSEPTQAISESDVQWRTAGDRIQTLLDSCAAGGAAAQER
MAV_2683|M.avium_104 ----MAHRPQE--------TQDDAQWRTAGDRIQTLLDAASAGGAVARER
: . . *:: : : . ::
Mvan_2369|M.vanbaalenii_PYR-1 AEELKAAVEEVHRAGLTTIVARMRADDAAR----PLLFDLVDDPVVRMLL
TH_1027|M.thermoresistible__bu ADEVRAAIEEVHRVGLTTIVRRMRADEGAK----QVLFELVDDPVVRMLL
MSMEG_2268|M.smegmatis_MC2_155 AEELKAALEEIHRVGLVTIVKRMRSGEATR----EALFELVDDPVVRMLF
MMAR_1868|M.marinum_M ASQLVREVVGLYGAGLERIMRLLSQPGLDPDAHIDLAERLATDDLVASLL
MAV_2683|M.avium_104 AEQLVSEVAGFYGAGLGRILDLIDDPAIP-----DILDRLVADDLVASLL
*.:: : .: .** *: : *. * :* *:
Mvan_2369|M.vanbaalenii_PYR-1 TLHGIIRPDPVTLAERALAAVRPQLHSHGGDVTLVRVDDR-----VAFVR
TH_1027|M.thermoresistible__bu SLHGIIRPDPVTQAHRALEAVRPQLHSHGGDVTLVRVEDG-----VAQVR
MSMEG_2268|M.smegmatis_MC2_155 TLHGIIRPDTTTLAEQALAQVRPQLRSHGGDVTLVRIESG-----TAFVR
MMAR_1868|M.marinum_M LVHGLHPHDVHRRVSDALDGVRPYLGSHGGDVELIDVVTGRSGDITVRLA
MAV_2683|M.avium_104 LVHGLHPHDVGRRVADALDSVRPYLGSHGGDVHLLEIVGD-----TVRLA
:**: * . ** *** * ****** *: : .. :
Mvan_2369|M.vanbaalenii_PYR-1 LEGACNGCSMASVTLRELVEAALLQGVPNLEAVEVVSGEATP------TL
TH_1027|M.thermoresistible__bu LEGACNGCSMSAVTLRDLVTGTLTEQVPAITRVEVLPNEPSP------TL
MSMEG_2268|M.smegmatis_MC2_155 LEGACNGCSMSAVTLRQLVETALLEGVQGVSKVEVLPNEPTP------TV
MMAR_1868|M.marinum_M FTGSCKSCPSSAVTLELAVQDAVRAAAPEISAIEVVAAEPAG---TAG-V
MAV_2683|M.avium_104 LDGSCKSCPSSAVTLELTVEDAVRSAAPEISSIEVVSEPVSAGTATAGVV
: *:*:.*. ::***. * :: . : :**:. : :
Mvan_2369|M.vanbaalenii_PYR-1 IPVEALRIPPRAE------PDGWVNLGSVDSIAVDEIRVL--------GG
TH_1027|M.thermoresistible__bu IPVEALRIGLDLR------DDGWVGAGPAADTSGDRVG------------
MSMEG_2268|M.smegmatis_MC2_155 ISIDAVLAGSTVRR---RAEEGWVRAGSVDAAAVDDVTPVTVTAPDGSEV
MMAR_1868|M.marinum_M ISAESLLSRVHSNGSNASPATSWHPVPELAELVPGEVAGFAIG-----DT
MAV_2683|M.avium_104 IQAESLLAKIHAKSGNP---TAWHQLPEIGELAPGQVGGFLVT-----DT
* ::: . .* . :
Mvan_2369|M.vanbaalenii_PYR-1 DVLVVNIGQKLSAFYNECAHEALPLGDAVFDATAG----TLTCPWHGFCY
TH_1027|M.thermoresistible__bu -VIIVNVDQRFAVYRNECAHEGLPLDNAVLDTADG----TLTCPWHGFCY
MSMEG_2268|M.smegmatis_MC2_155 RLLLVNLGQRMSAYRNECAHEALPLDGAVLDVGNG----TLTCPWHGFCY
MMAR_1868|M.marinum_M TLLACRVGEQVLAYRDHCPICRDSLAGAQLLG------TLLRCPGCRTDF
MAV_2683|M.avium_104 AVLACRVEDQLYVYRDHCPVCEQSLAGAALHRRMGAADVLLRCPRCHSHF
:: .: ::. .: :.*. .* .* : * ** :
Mvan_2369|M.vanbaalenii_PYR-1 DAESGECLSAPG------VQLEQLPLRIDDGDVWVRTGR-------
TH_1027|M.thermoresistible__bu DAASGECLSAPG------AQLEQLPLRIDRGELWVRVGP-------
MSMEG_2268|M.smegmatis_MC2_155 DATSGECMSAPG------VQLEPLPLRIDDGDIWVRVTG-------
MMAR_1868|M.marinum_M DVVQAGVPAGAG---HQELHLDPLPLLIRDGVPSIALRGQELGAIA
MAV_2683|M.avium_104 DVVHAGAGIDVDQDNHPEHHLDPIPLLVRDGVLSIAVPAEPLSTSA
*. . . :*: :** : * :