For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSETGAPLRVVILGSTGSIGTQALDVIAANPDRFQVVGLAAGGGNTDLLARQRAETGVTNIAVADEAAAT AVGGVTYSGPDAATRLVENTEADVVLNALVGALGLKPTLAALATGARLALANKESLVAGGPLVQKAAAPG QIVPVDSEHSALAQCLRGGTGDEVARLVLTASGGPFHGWTADRLEDVTPEQAAAHPTWSMGPMNTLNSAS LVNKGLELIETHLLFGVDYDRIDVVVHPQSIVHSMVTFTDGSTLAQASPPDMKLPIALALGWPSRVPGAA PACDFTTASRWDFEPLDDEVFPAVALAREAGRGGGCLTAVYNAANEEAAEAFLNGRIRFPAIVRTVARVL RAADQWAAEPATVDDVLDAQDWARDRARRAVEQEPAGSRR
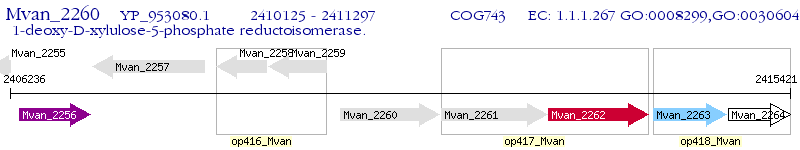
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2260 | - | - | 100% (390) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2895c | dxr | 1e-174 | 82.62% (374) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. gilvum PYR-GCK | Mflv_4083 | - | 0.0 | 91.62% (382) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. tuberculosis H37Rv | Rv2870c | dxr | 1e-174 | 82.62% (374) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. leprae Br4923 | MLBr_01583 | - | 1e-167 | 77.78% (387) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. abscessus ATCC 19977 | MAB_3171c | - | 1e-167 | 79.16% (379) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. marinum M | MMAR_1836 | dxr | 1e-175 | 80.63% (382) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase Dxr |
| M. avium 104 | MAV_3727 | dxr | 1e-176 | 81.05% (380) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. smegmatis MC2 155 | MSMEG_2578 | dxr | 1e-175 | 82.71% (376) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
| M. thermoresistible (build 8) | TH_1401 | - | 1e-138 | 85.61% (285) | PROBABLE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE DXR |
| M. ulcerans Agy99 | MUL_2085 | dxr | 1e-175 | 80.37% (382) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2260|M.vanbaalenii_PYR-1 ------MSETGAPLRVVILGSTGSIGTQALDVIAANPDRFQVVGLAAGGG
Mflv_4083|M.gilvum_PYR-GCK ------MNER---LRVLILGSTGSIGTQALEVIAANPDRFEVVGLAAGGG
TH_1401|M.thermoresistible__bu --------------------------------------------------
MSMEG_2578|M.smegmatis_MC2_155 MTTSA-ASGEPGRQRVLILGSTGSIGTQALEVIAANPDRFEVVGLAAGGG
MAB_3171c|M.abscessus_ATCC_199 --------MSSEVCRVLLLGSTGSIGTQALEVIAANPERFEVVGLAAGGG
Mb2895c|M.bovis_AF2122/97 MTNST-DGRADGRLRVVVLGSTGSIGTQALQVIADNPDRFEVVGLAAGGA
Rv2870c|M.tuberculosis_H37Rv MTNST-DGRADGRLRVVVLGSTGSIGTQALQVIADNPDRFEVVGLAAGGA
MLBr_01583|M.leprae_Br4923 MNNPI-EGHAGGRLRVLVLGSTGSIGTQALEVIAANPDRFEVVGLAAGGA
MMAR_1836|M.marinum_M MTTAT-DGSADGRLRVLVLGSTGSIGTQALEVIAANPDRFEVVGLAAGGS
MUL_2085|M.ulcerans_Agy99 MTTAT-DGFADGRLRVLVLGSTGSIGTQALEVIAANPDRFEVVGLAAGGS
MAV_3727|M.avium_104 MTNPTSDGQPRGRLRVLVLGSTGSIGTQALQVIAANPDRFEVVGLAAGGA
Mvan_2260|M.vanbaalenii_PYR-1 NTDLLARQRAETGVTNIAVADEAAATAVGGVTYSGPDAATRLVENTEADV
Mflv_4083|M.gilvum_PYR-GCK NRDLLARQRAETGVTNIAVADEAAAAALGEVTYSGPDAVTRLVEATEADV
TH_1401|M.thermoresistible__bu --------------------------------------------------
MSMEG_2578|M.smegmatis_MC2_155 NPELLAAQRAQTGVAAVAVADPAAAEAVGDVRYSGPDAVTRLVEDTEADV
MAB_3171c|M.abscessus_ATCC_199 NVELLRQQISETGVTNVAVADERAAVRLDIPVLSGPDAVTELVENTEADV
Mb2895c|M.bovis_AF2122/97 HLDTLLRQRAQTGVTNIAVADEHAAQRVGDIPYHGSDAATRLVEQTEADV
Rv2870c|M.tuberculosis_H37Rv HLDTLLRQRAQTGVTNIAVADEHAAQRVGDIPYHGSDAATRLVEQTEADV
MLBr_01583|M.leprae_Br4923 QLDTLLRQRAATGVTNIAIADDRAAQLAGDIPYHGTDAVTRLVEETEADV
MMAR_1836|M.marinum_M NLDTLLRQRAETGVTEIAIADERAAQLAGDIAYQGPDAVTRLVEETEADV
MUL_2085|M.ulcerans_Agy99 NLDTLRRQRAETGVTEIAIADERAAQLAGDIAYQEPDAVTRLVEETEADV
MAV_3727|M.avium_104 NLDTLLRQRAETGVTNVAVADEHAARRAGDIPFCGPEAATRLVEETEADV
Mvan_2260|M.vanbaalenii_PYR-1 VLNALVGALGLKPTLAALATGARLALANKESLVAGGPLVQKAAAPGQIVP
Mflv_4083|M.gilvum_PYR-GCK VLNGLVGALGLKPTLAALATGARLALANKESLVAGGPLVQKAAAPGQIVP
TH_1401|M.thermoresistible__bu --------LGLEPTLAALDTGARLALANKESLVAGGSLVLKAAGPGQIVP
MSMEG_2578|M.smegmatis_MC2_155 VLNALVGALGLQPTLAALATGARLALANKESLVAGGPLVLKAAAPGQIVP
MAB_3171c|M.abscessus_ATCC_199 VLNALVGALGLRPTLAALKSGARLALANKESLVAGGPLVTKAAAPGQIVP
Mb2895c|M.bovis_AF2122/97 VLNALVGALGLRPTLAALKTGARLALANKESLVAGGSLVLRAARPGQIVP
Rv2870c|M.tuberculosis_H37Rv VLNALVGALGLRPTLAALKTGARLALANKESLVAGGSLVLRAARPGQIVP
MLBr_01583|M.leprae_Br4923 VLNALVGALGLRPTLAALHTGARLALANKESLVAGGSLVLAAAQPGQIVP
MMAR_1836|M.marinum_M VLNALVGALGLRPTLAALASGARLALANKESLVAGGPLVLQAAQPGQIVP
MUL_2085|M.ulcerans_Agy99 VLNALVGALGLRPTLAALASGARLALANKESLVAGGPLVLQAAQPGQIVP
MAV_3727|M.avium_104 VLNALVGALGLRPTLAALESGARLALANKESLIAGGPLVLAAAAPGQIVP
***.****** :************:***.** ** ******
Mvan_2260|M.vanbaalenii_PYR-1 VDSEHSALAQCLRGGTGDEVARLVLTASGGPFHGWTADRLEDVTPEQAAA
Mflv_4083|M.gilvum_PYR-GCK VDSEHSALAQCLRGGSADEVATLVLTASGGPFRGWSVDRLEDVTPEQAGA
TH_1401|M.thermoresistible__bu VDSEHSAMAQCLRGGSPDEVARIVLTASGGPFRGWTAEQLETVTPEQAGA
MSMEG_2578|M.smegmatis_MC2_155 VDSEHSAMAQCLRGGTRAELDKIVLTASGGPFLGWSAEDLKSVTPEQAGK
MAB_3171c|M.abscessus_ATCC_199 VDSEHSALAQCLRGGTRGEVAQLILTASGGPFRGWTAQQLTAVTPEQAGA
Mb2895c|M.bovis_AF2122/97 VDSEHSALAQCLRGGTPDEVAKLVLTASGGPFRGWSAADLEHVTPEQAGA
Rv2870c|M.tuberculosis_H37Rv VDSEHSALAQCLRGGTPDEVAKLVLTASGGPFRGWSAADLEHVTPEQAGA
MLBr_01583|M.leprae_Br4923 VDSEHSALAQCLRGGTPDEVAKLVLTASGGPFRGWNAGDLERVTPEQAGV
MMAR_1836|M.marinum_M VDSEHSALAQCLRAGTPDEVAKLVLTASGGPFRGWSAADLQEVTPEQAGA
MUL_2085|M.ulcerans_Agy99 VDSEHSALAQCLRAGTPDEVAKLVLTASGGPFRGWSAADLQEVTPEQAGA
MAV_3727|M.avium_104 VDSEHSALAQCLRGGTGDEVAKLVLTASGGPFRGWTAEELEHVTPEQAGA
*******:*****.*: *: ::******** **.. * ******.
Mvan_2260|M.vanbaalenii_PYR-1 HPTWSMGPMNTLNSASLVNKGLELIETHLLFGVDYDRIDVVVHPQSIVHS
Mflv_4083|M.gilvum_PYR-GCK HPTWSMGPMNTLNSASLVNKGLELIETHLLFGIDYDRIDVVVHPQSIVHS
TH_1401|M.thermoresistible__bu HPTWSMGPMNTLNSASLVNKGLELIETHLLFGIDYDRIDVVVHPQSIVHS
MSMEG_2578|M.smegmatis_MC2_155 HPTWSMGPMNTLNSATLVNKGLELIETHLLFGVDYDDIDVVVHPQSIVHS
MAB_3171c|M.abscessus_ATCC_199 HPTWSMGPMNTLNSASLVNKGLELIETHLLFGVPYRQIGVVVHPQSIVHS
Mb2895c|M.bovis_AF2122/97 HPTWSMGPMNTLNSASLVNKGLEVIETHLLFGIPYDRIDVVVHPQSIIHS
Rv2870c|M.tuberculosis_H37Rv HPTWSMGPMNTLNSASLVNKGLEVIETHLLFGIPYDRIDVVVHPQSIIHS
MLBr_01583|M.leprae_Br4923 HPTWSMGTMNTLNSASLVNKGLELIEANLLFGIPYDRIEVVVHPQSIVHS
MMAR_1836|M.marinum_M HPTWSMGPMNTLNSASLVNKGLELIETHLLFGIPYERIEVVVHPQSIVHS
MUL_2085|M.ulcerans_Agy99 HPTWSMGPMNTLNSASLVNKGLELIETHLLFGISYERIEVVVHPQSIVHS
MAV_3727|M.avium_104 HPTWSMGPMNTLNSASLVNKGLELIETHLLFGIPYDRIEVVVHPQSIVHS
*******.*******:*******:**::****: * * ********:**
Mvan_2260|M.vanbaalenii_PYR-1 MVTFTDGSTLAQASPPDMKLPIALALGWPSRVPGAAPACDFTTASRWDFE
Mflv_4083|M.gilvum_PYR-GCK MVTFTDGSTLAQASPPDMKLPIALALGWPARVAGAAAACDFSTASRWDFE
TH_1401|M.thermoresistible__bu MVTFTDGSTLAQASPPDMKLPIALALSWPDRVPGAAPACDFSTASTWTFE
MSMEG_2578|M.smegmatis_MC2_155 MATFTDGSTLAQASPPDMKLPIALALGWPDRIAGAAAACDFSTASTWEFL
MAB_3171c|M.abscessus_ATCC_199 MVTFTDGSTLAQASPPDMKLPIALALGWPDRVPAAASACDFSTASTWEFE
Mb2895c|M.bovis_AF2122/97 MVTFIDGSTIAQASPPDMKLPISLALGWPRRVSGAAAACDFHTASSWEFE
Rv2870c|M.tuberculosis_H37Rv MVTFIDGSTIAQASPPDMKLPISLALGWPRRVSGAAAACDFHTASSWEFE
MLBr_01583|M.leprae_Br4923 MVTFIDGSTIAQASPPDMKLPISLALGWPQRVGGAARACAFTTASTWEFE
MMAR_1836|M.marinum_M MVTFVDGSTIAQASPPDMKLPISLALGWPRRVPGAARYCDFSQAQTWEFE
MUL_2085|M.ulcerans_Agy99 MVTFVDGSTIAQASPPDMKLPISLALGWPRRVPGAARYCDFSQAQTWEFE
MAV_3727|M.avium_104 MVTFVDGSTLAQASPPDMRLPISLALGWPHRVPGAAACCDFSTASSWEFE
*.** ****:********:***:***.** *: .** * * *. * *
Mvan_2260|M.vanbaalenii_PYR-1 PLDDEVFPAVALAREAGRGGGCLTAVYNAANEEAAEAFLNGRIRFPAIVR
Mflv_4083|M.gilvum_PYR-GCK PLDDEVFPAVALAREAGKTGGCMTAIYNAANEEAAEAFLAGRIRFPAIVR
TH_1401|M.thermoresistible__bu PVDNEVFPAVELARQAGERGGCLTAVYNAANEEAAAAFLAGRIRFPAIVR
MSMEG_2578|M.smegmatis_MC2_155 PLDNAVFPAVDLARFAGKQGGCLTAVYNSANEEAAEAFLDGRIGFPDIVE
MAB_3171c|M.abscessus_ATCC_199 PLDAQVFPAVDLARSAGEAGGCMTAVYNAANETAAAAFLDGRIGFRSIVR
Mb2895c|M.bovis_AF2122/97 PLDTDVFPAVELARQAGVAGGCMTAVYNAANEEAAAAFLAGRIGFPAIVG
Rv2870c|M.tuberculosis_H37Rv PLDTDVFPAVELARQAGVAGGCMTAVYNAANEEAAAAFLAGRIGFPAIVG
MLBr_01583|M.leprae_Br4923 PLDIDVFPAVELARHAGQIGGCMTAIYDAANEEAAEAFLQGRIGFPAIVA
MMAR_1836|M.marinum_M PLDDEVFPAVELARHAGETGGCMTAVYNAANEEAAAAFLAGRISFPAIVG
MUL_2085|M.ulcerans_Agy99 PLDDEVFPAVELARHAGETGGCMTAVYNAANEEAAAAFLAGRISFPAIVG
MAV_3727|M.avium_104 PLDNEVFPAVELARHAGQAGGCMTAIYNAANEEAAAAFLAGRVSFGAIVE
*:* ***** *** ** ***:**:*::*** ** *** **: * **
Mvan_2260|M.vanbaalenii_PYR-1 TVARVLRAADQWA----AEPATVDDVLDAQDWARDRARR------AVEQE
Mflv_4083|M.gilvum_PYR-GCK TVAEVLRAADQWA----AEPATVDDVLDAQNWARDRARS------AVERE
TH_1401|M.thermoresistible__bu IVAEVLRAADAWA----AEPATVDDVLEAQRWARTVARH------AVDRS
MSMEG_2578|M.smegmatis_MC2_155 TVGDVLHAADRWA----AEPATVDDVLDAQRWAREQARG------VVEQK
MAB_3171c|M.abscessus_ATCC_199 TIADVLDVASQWEGT-SAEPATVDDVLDAQRWAGQQAAQ------FIFAE
Mb2895c|M.bovis_AF2122/97 IIADVLHAADQWA----VEPATVDDVLDAQRWARERAQRAVSGMASVAIA
Rv2870c|M.tuberculosis_H37Rv IIADVLHAADQWA----VEPATVDDVLDAQRWARERAQRAVSGMASVAIA
MLBr_01583|M.leprae_Br4923 TIADVLQRADQWAPQWGEGPATVDDVLDAQRWARERA------LCAVATA
MMAR_1836|M.marinum_M TIADVLHAADQWA----VSPANVDDVLDAQRWARQRAQR------AIAKA
MUL_2085|M.ulcerans_Agy99 TIADVLHAADQWA----VSPANVDDVLDAQRWARQRAQR------AIAKA
MAV_3727|M.avium_104 TIADVLHAADQWAPKFSETPANVDDVLDAQRWARQQAQR------AVAQA
:. ** *. * **.*****:** ** * :
Mvan_2260|M.vanbaalenii_PYR-1 PAGS-------RR-----
Mflv_4083|M.gilvum_PYR-GCK SAGS-------RS-----
TH_1401|M.thermoresistible__bu LDGK-------GLLTT--
MSMEG_2578|M.smegmatis_MC2_155 SVRR-------GLVTK--
MAB_3171c|M.abscessus_ATCC_199 ENRT--------------
Mb2895c|M.bovis_AF2122/97 STAKPGAAGRHASTLERS
Rv2870c|M.tuberculosis_H37Rv STAKPGAAGRHASTLERS
MLBr_01583|M.leprae_Br4923 SSGK-----VSDMVLERS
MMAR_1836|M.marinum_M SPAG-----ACEKVSGLA
MUL_2085|M.ulcerans_Agy99 SPAG-----ACEKVSGLA
MAV_3727|M.avium_104 RPAT-----VSAKTPGVV