For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKLITAIVKPFTLEDVKTGLEQTGILGMTVSEVQGYGRQKGHTEVYRGAEYSVDFVPKVRVEVVVDDAAV DKVVDVIVQAARTGKIGDGKVWVSPVETVVRVRTGERGADAL
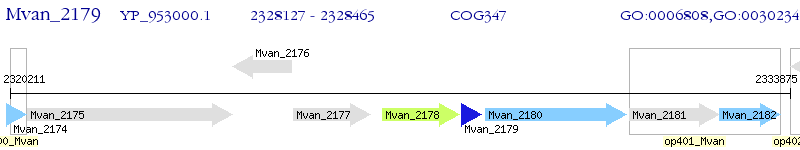
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2179 | - | - | 100% (112) | nitrogen regulatory protein P-II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2943c | glnB | 2e-53 | 87.50% (112) | nitrogen regulatory protein P-II GLNB |
| M. gilvum PYR-GCK | Mflv_4184 | - | 3e-59 | 100.00% (112) | nitrogen regulatory protein P-II |
| M. tuberculosis H37Rv | Rv2919c | glnB | 2e-53 | 87.50% (112) | nitrogen regulatory protein P-II GLNB |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3239c | - | 3e-56 | 93.75% (112) | nitrogen regulatory protein P-II |
| M. marinum M | MMAR_1788 | glnB | 1e-53 | 87.50% (112) | nitrogen regulatory protein P-II GlnB |
| M. avium 104 | MAV_3774 | - | 5e-54 | 88.39% (112) | nitrogen regulatory protein P-II |
| M. smegmatis MC2 155 | MSMEG_2426 | - | 9e-59 | 99.11% (112) | nitrogen regulatory protein P-II |
| M. thermoresistible (build 8) | TH_1480 | glnB | 5e-58 | 96.43% (112) | PROBABLE NITROGEN REGULATORY PROTEIN P-II GLNB |
| M. ulcerans Agy99 | MUL_2036 | glnB | 2e-53 | 86.61% (112) | nitrogen regulatory protein P-II GlnB |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2943c|M.bovis_AF2122/97 MKLITAIVKPFTLDDVKTSLEDAGVLGMTVSEIQGYGRQKGHTEVYRGAE
Rv2919c|M.tuberculosis_H37Rv MKLITAIVKPFTLDDVKTSLEDAGVLGMTVSEIQGYGRQKGHTEVYRGAE
MAV_3774|M.avium_104 MKLITAIVKPFTLDDVKTSLEDAGVLGMTVSEIQGYGRQKGHTEVYRGAE
MMAR_1788|M.marinum_M MKLITAIVKPFTLDDVKTSLEEAGVLGMTVSEIQGYGRQKGHTEVYRGAE
MUL_2036|M.ulcerans_Agy99 MKLITAIVKPFTLDDVKTSLEEAGVLGMTVSEIQGYGRQKGHTEVYRGAE
Mvan_2179|M.vanbaalenii_PYR-1 MKLITAIVKPFTLEDVKTGLEQTGILGMTVSEVQGYGRQKGHTEVYRGAE
Mflv_4184|M.gilvum_PYR-GCK MKLITAIVKPFTLEDVKTGLEQTGILGMTVSEVQGYGRQKGHTEVYRGAE
TH_1480|M.thermoresistible__bu MKLITAIIKPFTLEDVKTGLEQTGILGMTVSEVQGYGRQKGHTEVYRGAE
MSMEG_2426|M.smegmatis_MC2_155 MKLITAIVKPFTLEDVKTGLEQTGILGMTVSEVQGYGRQKGHTEVYRGAE
MAB_3239c|M.abscessus_ATCC_199 MKLVTAIVKPFTLEDVKTGLEQAGILGMTVSEVQGYGRQKGHTEVYRGAE
***:***:*****:****.**::*:*******:*****************
Mb2943c|M.bovis_AF2122/97 YSVDFVPKVRIEVVVDDSIVDKVVDSIVRAARTGKIGDGKVWVSPVDTIV
Rv2919c|M.tuberculosis_H37Rv YSVDFVPKVRIEVVVDDSIVDKVVDSIVRAARTGKIGDGKVWVSPVDTIV
MAV_3774|M.avium_104 YSVDFVPKVRIEVVVDDSIVDKVVDSIVRAARTGKIGDGKVWVSPVETIV
MMAR_1788|M.marinum_M YSVDFVPKVRIEVVVDDSIVDKVVDSIVRAARTGKIGDGKVWVSPVDTIV
MUL_2036|M.ulcerans_Agy99 YSVDFVPKVRIEVVVDDSIVDKVVDSIVRVARTGKIGDGKVWVSPVDTIV
Mvan_2179|M.vanbaalenii_PYR-1 YSVDFVPKVRVEVVVDDAAVDKVVDVIVQAARTGKIGDGKVWVSPVETVV
Mflv_4184|M.gilvum_PYR-GCK YSVDFVPKVRVEVVVDDAAVDKVVDVIVQAARTGKIGDGKVWVSPVETVV
TH_1480|M.thermoresistible__bu YSVDFVPKVRVEVLVEDSAVDKVVDVIVQAARTGKIGDGKVWVSPVETVV
MSMEG_2426|M.smegmatis_MC2_155 YSVDFVPKVRVEVVVDDSAVDKVVDVIVQAARTGKIGDGKVWVSPVETVV
MAB_3239c|M.abscessus_ATCC_199 YSVDFVPKVRVEVVVDDSAVDKVVEVIVTAARTGKIGDGKVWVSPVDSVV
**********:**:*:*: *****: ** .****************:::*
Mb2943c|M.bovis_AF2122/97 RVRTGERGHDAL
Rv2919c|M.tuberculosis_H37Rv RVRTGERGHDAL
MAV_3774|M.avium_104 RVRTGERGTDAL
MMAR_1788|M.marinum_M RVRTGERGPDAL
MUL_2036|M.ulcerans_Agy99 RVRTGERGPDAL
Mvan_2179|M.vanbaalenii_PYR-1 RVRTGERGADAL
Mflv_4184|M.gilvum_PYR-GCK RVRTGERGADAL
TH_1480|M.thermoresistible__bu RVRTGERGADAL
MSMEG_2426|M.smegmatis_MC2_155 RVRTGERGADAL
MAB_3239c|M.abscessus_ATCC_199 RVRTGERGADAL
******** ***