For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDPDPLVPRPAATVMLVRDTPEGIKVFLMRRHSAMDFVAGVMVFPGGGVDDRDRNADVAWHGPDRQWWA ERFGVDVELAEALVCAAARETFEESGVLFAGAADDPDLLVDDASVYRDQRAALENKSLSFGEFLRSEKLM LRADLLRPWANWVTPKEERTRRYDTYFFVGALPQGQRADGDNTETDQAGWVTPQAALDDFADGRSFLLPP TWTQLDALNGRTVAEVLAVERKIVAVEPSLAAQKGGNWEIEFFNSDRYNEARNRRSPQGYADGVPLT
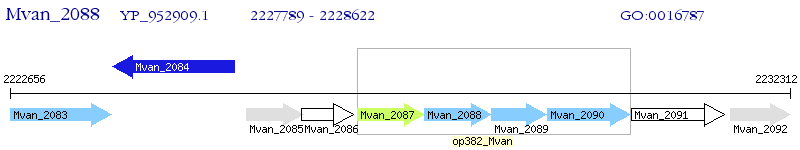
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2088 | - | - | 100% (277) | NUDIX hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3066c | - | 1e-107 | 68.95% (277) | hypothetical protein Mb3066c |
| M. gilvum PYR-GCK | Mflv_4271 | - | 1e-153 | 94.18% (275) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv3040c | - | 1e-107 | 68.95% (277) | hypothetical protein Rv3040c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3383c | - | 2e-84 | 59.47% (264) | hypothetical protein MAB_3383c |
| M. marinum M | MMAR_1661 | - | 1e-110 | 73.31% (266) | hypothetical protein MMAR_1661 |
| M. avium 104 | MAV_3905 | - | 1e-112 | 72.76% (268) | hydrolase, nudix family protein, putative |
| M. smegmatis MC2 155 | MSMEG_2327 | - | 1e-115 | 75.46% (273) | nudix hydrolase |
| M. thermoresistible (build 8) | TH_1314 | - | 1e-123 | 80.90% (267) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1898 | - | 1e-110 | 73.31% (266) | hypothetical protein MUL_1898 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3066c|M.bovis_AF2122/97 ---MNSPREPLVPPPTPRPAATVMLVRDPDAGSASGLAVFLMRRHAAMDF
Rv3040c|M.tuberculosis_H37Rv ---MNSPREPLVPPPTPRPAATVMLVRDPDAGSASGLAVFLMRRHAAMDF
MMAR_1661|M.marinum_M ---MNSPPDS--VPLQARPAATVMLVR--DTGLDTGLSVFLMRRHAAMDF
MUL_1898|M.ulcerans_Agy99 ---MNSPPDP--VPLQARPAATVMLVR--DTGLDTGLSVFLMRRHAAMDF
MAV_3905|M.avium_104 ---MTSPEEA---PLVPRPAATVMLVR----EAPTGLKVFLMRRHSRMEF
Mvan_2088|M.vanbaalenii_PYR-1 ------MTDP--DPLVPRPAATVMLVRDT-PEG---IKVFLMRRHSAMDF
Mflv_4271|M.gilvum_PYR-GCK MSELKTTTEP--DPLVPRPAATVMLVRDT-PEG---IKIFLMRRHSAMDF
TH_1314|M.thermoresistible__bu ---MTRSEPP--APLPPRPAATVMLIRDR-HDGAAGLEVFLMRRHSAMDF
MSMEG_2327|M.smegmatis_MC2_155 -------MTD--DPLVPLPAATVMLIRDVHREGRADLEVFLMRRHAAMDF
MAB_3383c|M.abscessus_ATCC_199 ---MTAATPP-----EPKPAATVVLIR----DGAVGVEAFLMRRDNAMAF
. *****:*:* : *****. * *
Mb3066c|M.bovis_AF2122/97 AAGVMVFPGGGVDDRDRDADLGRLGAWAGPPPQWWAQRFGIEPDLAEALV
Rv3040c|M.tuberculosis_H37Rv AAGVMVFPGGGVDDRDRDADLGRLGAWAGPPPQWWAQRFGIEPDLAEALV
MMAR_1661|M.marinum_M AAGVMVFPGGGVDDRDRNADLGGLGAWAGPPPLWWAQRFGIQPDLAEALV
MUL_1898|M.ulcerans_Agy99 AAGVMVFPGGGVDDRDRNADLGGLGAWAGPPPLWWAQRFGIQPDLAEALV
MAV_3905|M.avium_104 AAGVMVFPGGGVDERDRNADLGGLGAWAGPPPQWWAQRFGIEPDLAEALV
Mvan_2088|M.vanbaalenii_PYR-1 VAGVMVFPGGGVDDRDRNADV----AWHGPDRQWWAERFGVDVELAEALV
Mflv_4271|M.gilvum_PYR-GCK VAGVMVFPGGGVDDRDRNADV----AWHGPDRHWWAGRLGVDAELAEALV
TH_1314|M.thermoresistible__bu VAGVMVFPGGGVDDRDRNADI----AWMGPEPAWWAERFGVDEGLAEALV
MSMEG_2327|M.smegmatis_MC2_155 VAGVMVFPGGGVDDRDRNADI----AWYGPEPSWWAQRLGVDEGLAEALV
MAB_3383c|M.abscessus_ATCC_199 AGGMTVFPGGGVDSRDMHADV----PWLGPDVDWWAQQFGVTPELASALV
..*: ********.** .**: .* ** *** ::*: **.***
Mb3066c|M.bovis_AF2122/97 CAAARETFEESGVLFAGPVDQDHSAPNSIVSDASVYGDARRALADRTLSF
Rv3040c|M.tuberculosis_H37Rv CAAARETFEESGVLFAGPVDQDHSAPNSIVSDASVYGDARRALADRTLSF
MMAR_1661|M.marinum_M CAAARETFEESGVLFAGPADD----PDGIVGDASIYSESRAALADHTLSF
MUL_1898|M.ulcerans_Agy99 CAAARETFEESGVLFAGPADD----PDGIVGDASIYSESRAALADHTLSF
MAV_3905|M.avium_104 CAAARETFEESGVLFAGPAGA----PDSIVGDASVYRDARRALADGTLSF
Mvan_2088|M.vanbaalenii_PYR-1 CAAARETFEESGVLFAGAADDPD----LLVDDASVYRDQRAALENKSLSF
Mflv_4271|M.gilvum_PYR-GCK CAAARETFEESGVLFAGAADDPD----LLVDDASVYRDQRAALENKSLSF
TH_1314|M.thermoresistible__bu CAAARETFEECGVLFAGAADDSD----VLVDDASVYHDARRALADKSLSF
MSMEG_2327|M.smegmatis_MC2_155 CAAARETFEESGVLFAGPQNGPGD--PGIIADASVYGDARAALANHSLSF
MAB_3383c|M.abscessus_ATCC_199 CAAARETFEECGVLFAGTSEN------SIVEDAAVYHQARKDLSDKTLSF
**********.******. :: **::* : * * : :***
Mb3066c|M.bovis_AF2122/97 ADFLQREKLVLRSDLLRPWANWVTPEAELTRRYDTYFFVGALPEGQRADG
Rv3040c|M.tuberculosis_H37Rv ADFLQREKLVLRSDLLRPWANWVTPEAELTRRYDTYFFVGALPEGQRADG
MMAR_1661|M.marinum_M ADFLRREDLVLRADLLRPWANWITPEAERTRRYDTFFFVGALPEGQRADG
MUL_1898|M.ulcerans_Agy99 ADFLRRENLVLRADLLRPWANWITPEAERTRRYDTFFFVGALPEGQRADG
MAV_3905|M.avium_104 ADFLRTEKLELRSDLLRPWANWVTPEAERTRRYDTYFFVGALPQGQRADG
Mvan_2088|M.vanbaalenii_PYR-1 GEFLRSEKLMLRADLLRPWANWVTPKEERTRRYDTYFFVGALPQGQRADG
Mflv_4271|M.gilvum_PYR-GCK GEFLRSEKLMLRADLLRPWANWVTPKEERTRRYDTYFFVGALPQGQRADG
TH_1314|M.thermoresistible__bu GEFLRAENLVLRADLLRPWANWVTPEEERTRRYDTYFFVGALPGGQRADG
MSMEG_2327|M.smegmatis_MC2_155 ADFLRDEKLVLRADLLRPWANWVTPKEERTRRYDTYFFVAALPEGQRADG
MAB_3383c|M.abscessus_ATCC_199 AEFLQREGLQLRADLLRPWANWITPEAE-PRRYDTFFFVAALPAGQDADG
.:**: * * **:*********:**: * .*****:***.*** ** ***
Mb3066c|M.bovis_AF2122/97 ENTESDRAGWVLPADAIADFAAGRNFLLPPTWTQLDSLA--GHTVADVLA
Rv3040c|M.tuberculosis_H37Rv ENTESDRAGWVLPADAIADFAAGRNFLLPPTWTQLDSLA--GHTVADVLA
MMAR_1661|M.marinum_M ENTESDRAEWMLPRDAIADFAAGRNFLLPPTWTQLDALA--GHTVADVLA
MUL_1898|M.ulcerans_Agy99 ENTESDRAEWMLPRDAIADFAAGRNFLLPPTWTQLDALA--GHTVADVLA
MAV_3905|M.avium_104 QNTESDRAGWTTPEAALEDFSAGRTFLLPPTWTQLDSLA--GRTVADVLA
Mvan_2088|M.vanbaalenii_PYR-1 DNTETDQAGWVTPQAALDDFADGRSFLLPPTWTQLDALN--GRTVAEVLA
Mflv_4271|M.gilvum_PYR-GCK DNTETDKADWVTPQAALDDFAEGRSFLLPPTWTQLDALN--GRTVAEVLA
TH_1314|M.thermoresistible__bu DNTETDQADWVTPEQALADFAAGRTFLLPPTWTQLDSLA--GRTVAEVMA
MSMEG_2327|M.smegmatis_MC2_155 ENTETDRAFWCTPKAGLDDFEAGNTFLLPPTWTQLDSLD--GRTVAEVLA
MAB_3383c|M.abscessus_ATCC_199 NNTEAVASGWQTPAAAIAAFTERRSFLLPPTWSQLDAVSSAGASVAEVLG
:***: : * * .: * ..*******:***:: * :**:*:.
Mb3066c|M.bovis_AF2122/97 VERQIVPVQPQLA-RNGDNWEIEFFDSDRYNQARRSGGSTGWPL-----
Rv3040c|M.tuberculosis_H37Rv VERQIVPVQPQLA-RNGDNWEIEFFDSDRYNQARRSGGSTGWPL-----
MMAR_1661|M.marinum_M VERKIVPVQPNLA-LKGDNWEIEFFDSDRYNQARISGGSTVWPL-----
MUL_1898|M.ulcerans_Agy99 VERKIVPVQPNLA-LKGDNREIEFFDSDRYNQARISGGSTVWPL-----
MAV_3905|M.avium_104 VQRQIAPVQPHVE-IRGDNWVFEFFDSDRYHRAREAGG-LGWRH-----
Mvan_2088|M.vanbaalenii_PYR-1 VERKIVAVEPSLAAQKGGNWEIEFFNSDRYNEARNRRSPQGYADGVPLT
Mflv_4271|M.gilvum_PYR-GCK VERKIVAVEPSLAATKGGNWEIEFFDGDRYNAARNRRAPQGYTDGAPLA
TH_1314|M.thermoresistible__bu VERQIVAVEPHLRVDNG-NWEIEFFDSERYNAARNHRSP----------
MSMEG_2327|M.smegmatis_MC2_155 VERKIVTIEPSLS-TEGNNWEIEYFDSGRYNAARKHRSPGQ--------
MAB_3383c|M.abscessus_ATCC_199 VARSIAPIMPILT-EQDGRWFVQFDDVDRYEAARQGAVEIPEVTGPELS
* *.*..: * : .. . .:: : **. **