For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARLMGVDLPRDKRMEIALTYIYGIGRTRSQEILEATGISRDMRTKDLTDDQVTQLRDYIEGNLKVEGDL RREVQADIRRKIEIGCYQGLRHRRGLPVRGQRTKTNARTRKGPKRTIAGKKKAR
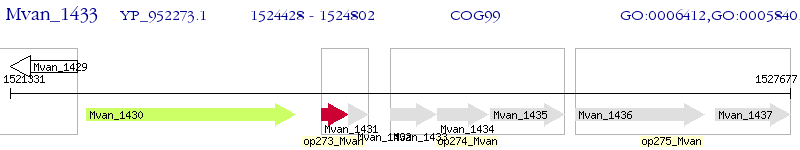
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1433 | rpsM | - | 100% (124) | 30S ribosomal protein S13 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3489c | rpsM | 2e-60 | 87.90% (124) | 30S ribosomal protein S13 |
| M. gilvum PYR-GCK | Mflv_4973 | rpsM | 3e-66 | 97.58% (124) | 30S ribosomal protein S13 |
| M. tuberculosis H37Rv | Rv3460c | rpsM | 2e-60 | 87.90% (124) | 30S ribosomal protein S13 |
| M. leprae Br4923 | MLBr_01960 | rpsM | 4e-63 | 91.13% (124) | 30S ribosomal protein S13 |
| M. abscessus ATCC 19977 | MAB_3773c | rpsM | 1e-61 | 91.94% (124) | 30S ribosomal protein S13 |
| M. marinum M | MMAR_1087 | rpsM | 6e-62 | 90.32% (124) | 30S ribosomal protein S13, RpsM |
| M. avium 104 | MAV_4401 | rpsM | 6e-55 | 91.82% (110) | 30S ribosomal protein S13 |
| M. smegmatis MC2 155 | MSMEG_1521 | rpsM | 2e-63 | 94.35% (124) | 30S ribosomal protein S13 |
| M. thermoresistible (build 8) | TH_0029 | rpsM | 5e-61 | 93.33% (120) | PROBABLE 30S RIBOSOMAL PROTEIN S13 RPSM |
| M. ulcerans Agy99 | MUL_0845 | rpsM | 4e-62 | 90.32% (124) | 30S ribosomal protein S13 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1087|M.marinum_M MARLVGVDLPRDKRMEIALTYIYGVGRTRSNEILAATGIDRNLRTRDLTD
MUL_0845|M.ulcerans_Agy99 MARLVGVDLPRDKRMEIALTYIYGVGRTRSNEILAATGIDRNLRTRDLTD
MAV_4401|M.avium_104 --------------MEIALTYIYGVGRTRSNEILAATGIDRDLRTRDLTD
Mb3489c|M.bovis_AF2122/97 MARLVGVDLPRDKRMEVALTYIFGIGRTRSNEILAATGIDRDLRTRDLTE
Rv3460c|M.tuberculosis_H37Rv MARLVGVDLPRDKRMEVALTYIFGIGRTRSNEILAATGIDRDLRTRDLTE
MLBr_01960|M.leprae_Br4923 MARLVGVDLPRDKRMEVALTYIYGIGRTRANEILEATGIERDLRTRDLTD
Mvan_1433|M.vanbaalenii_PYR-1 MARLMGVDLPRDKRMEIALTYIYGIGRTRSQEILEATGISRDMRTKDLTD
Mflv_4973|M.gilvum_PYR-GCK MARLMGVDLPRDKRMEIALTYIYGVGRTRSQEILEATGISRDQRTKDLTD
MAB_3773c|M.abscessus_ATCC_199 MARLVGVDLPRDKRMEIALTYIYGIGRTRSKEILAATGISPEQRSKDLTD
MSMEG_1521|M.smegmatis_MC2_155 MARLVGVDLPRDKRMEIALTYIYGIGRTRSNEILAATGIDKNMRTKDLTD
TH_0029|M.thermoresistible__bu ----VGVDLPRDKRMEIALTYIYGIGRTRSREILAATGIDKDMRTKDLTD
**:*****:*:****:.*** ****. : *::***:
MMAR_1087|M.marinum_M DQLTHLRDYIEANLKIEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
MUL_0845|M.ulcerans_Agy99 DQLTHLRDYIEANLKIEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
MAV_4401|M.avium_104 DQLTHLRDYIEANLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
Mb3489c|M.bovis_AF2122/97 EQLIHLRDYIEANLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGMPVRG
Rv3460c|M.tuberculosis_H37Rv EQLIHLRDYIEANLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGMPVRG
MLBr_01960|M.leprae_Br4923 DQLTHLRDYIEANLKVEGDLRREVQADIRRKMEIGCYQGLRHRRGLPVRG
Mvan_1433|M.vanbaalenii_PYR-1 DQVTQLRDYIEGNLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
Mflv_4973|M.gilvum_PYR-GCK DQVSQLRDYIEGNLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
MAB_3773c|M.abscessus_ATCC_199 DQVTQLREYIEATLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
MSMEG_1521|M.smegmatis_MC2_155 DQVTVLRDYIEGNLKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
TH_0029|M.thermoresistible__bu DQVTKLRDYIEENFKVEGDLRREVQADIRRKIEIGCYQGLRHRRGLPVRG
:*: **:*** .:*:***************:*************:****
MMAR_1087|M.marinum_M QRTKTNARTRKGPKRTIAGKKKAR
MUL_0845|M.ulcerans_Agy99 QRTKTNARTRKGPKRTIAGKKKAR
MAV_4401|M.avium_104 QRTKTNARTRKGPKRTIAGKKKAR
Mb3489c|M.bovis_AF2122/97 QRTKTNARTRKGPKRTIAGKKKAR
Rv3460c|M.tuberculosis_H37Rv QRTKTNARTRKGPKRTIAGKKKAR
MLBr_01960|M.leprae_Br4923 QRTKTNARTRKGPKRTIAGKKKAR
Mvan_1433|M.vanbaalenii_PYR-1 QRTKTNARTRKGPKRTIAGKKKAR
Mflv_4973|M.gilvum_PYR-GCK QRTKTNARTRKGPKRTIAGKKKAR
MAB_3773c|M.abscessus_ATCC_199 QRTKTNARTRKGPKRTIAGKKKAR
MSMEG_1521|M.smegmatis_MC2_155 QRTKTNARTRKGPKRTIAGKKKAR
TH_0029|M.thermoresistible__bu QRTKTNARTRKGPKRTIAGKKKAR
************************